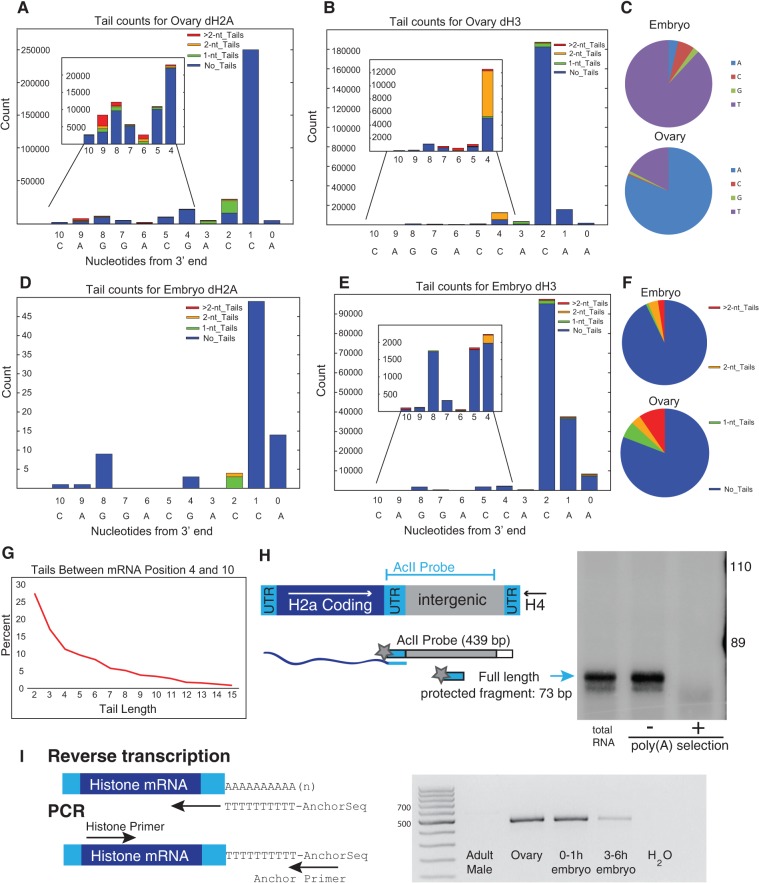
FIGURE 4.
Analysis of Drosophila histone mRNAs. (A,B) RNA from Drosophila ovary dH2a (A) and dH3 (B) was analyzed by EnD-Seq using a strategy to amplify the histone mRNAs selectively. Essentially all the 3′ ends mapped to the 3′ side of the stem in the stem–loop. The graphs represent the distribution of all recovered 3′ ends (with or without nontemplated tails) for each developmental stage. The inset shows the expansion of the distribution of 3′ ends from nucleotides −4 to −10 covering the 3′ side of the stem. (C) Pie charts showing nucleotide composition of single nucleotide tails in embryo (top) and ovary (bottom). (D,E) Distribution of 3′ ends and nontemplated tails in 3–6 h Drosophila embryos for (D) dH2a and (E) dH3. Note that the pattern of tail addition matches the ovary genes in (A) and (B) but the nucleotide composition is different. (F) Distribution of tail lengths for tails longer than 2 nt in the ovary RNA mapping between nucleotides 4 and 10 in the 3′ end of the stem. The tails >2 nt in the ovary were predominantly oligo(A). The tails of 1 or 2 nt in the embryo at nucleotides 4–10 were predominantly U's, and there were very few tails longer than 2 nt. (G) Chart showing the distribution of tail length between mRNA positions −4 and −10. Note that there are very few long tails. (H) S1 nuclease mapping of ovary histone mRNAs. Total RNA from ovaries was fractionated into poly(A+) and poly(A−) fractions on oligo(dT) cellulose. Total RNA, and equal proportions of the poly(A−) and poly(A+) RNA, were subjected to S1 nuclease mapping using the histone H2a gene labeled at the 3′ end of the AclI site as a probe. The S1 resistant fragments were resolved on a 6% polyacrylamine-7M urea gel and detected by autoradiography. A diagram of the S1 assay is shown on the left. The arrow indicates the fragment protected by the full-length mRNA. (I) RT-PCR analysis of the histone H2a mRNA. cDNA primed with oligo(dT) fused to an anchor primer was synthesized from 1 μg of total RNA from adult males, ovaries, 0–1 h embryos, and 3–6 h embryos, and then amplified using a primer near the 5′ end of the H2a mRNA. An amplicon of full-length H2a mRNA would be ∼500 nt long. Several amplicons were cloned and the majority had A-tails added in the 3′ end of the stem, consistent with the high-throughput sequencing data.