Abstract
Tuberculosis (TB) remains a global health problem, with vaccination being a necessary strategy for disease containment and elimination. A TB vaccine should be safe and immunogenic as well as efficacious in all affected populations, including HIV-infected individuals. We investigated the induction and maintenance of vaccine-induced memory CD4+ T cells following vaccination with the subunit vaccine H1/IC31. H1/IC31 was inoculated twice on study days 0 and 56 among HIV-infected adults with CD4+ lymphocyte counts of >350 cells/mm3. Whole venous blood stimulation was conducted with the H1 protein, and memory CD4+ T cells were analyzed using intracellular cytokine staining and polychromatic flow cytometry. We identified high responders, intermediate responders, and nonresponders based on detection of interleukin-2 (IL-2), tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ) expressing central (TCM) and effector memory CD4+ T cells (TEM) 182 days after the first immunization. Amplicon-based transcript quantification using next-generation sequencing was performed to identify differentially expressed genes that correlated with vaccine-induced immune responses. Genes implicated in resolution of inflammation discriminated the responders from the nonresponders 3 days after the first inoculation. The volunteers with higher expression levels of genes involved in antiviral innate immunity at baseline showed impaired H1-specific TCM and TEM maintenance 6 months after vaccination. Our study showed that in HIV-infected volunteers, expression levels of genes involved in the antiviral innate immune response affected long-term maintenance of H1/IC31 vaccine-induced cellular immunity. (The clinical trial was registered in the Pan African Clinical Trials Registry [PACTR] with the identifier PACTR201105000289276.)
INTRODUCTION
Vaccination represents one of the most successful health interventions for disease containment, elimination, and eventual eradication (1). Despite more than 50 years of widespread vaccination with Mycobacterium bovis bacille Calmette-Guérin (BCG), tuberculosis (TB) remains one of the world's most serious infectious diseases. In 2012, the World Health Organization (WHO) estimated that there were 9 million new clinical TB cases and 1 million people died from TB (2). Infection with HIV impairs host resistance, leading to a faster and higher rate of progression from latent to clinical TB (3). Consequently, there is an urgent need for development of improved TB vaccines that are more efficacious, safer, and more immunogenic than BCG in all populations, including HIV-infected individuals. H1/IC31 is a protein subunit vaccine against Mycobacterium tuberculosis, consisting of a fusion protein of the mycobacterial antigen 85B (Ag85B) and early secretory antigenic target 6 (ESAT-6) called hybrid 1 (H1) in combination with the adjuvant IC31. IC31 is composed of the cationic polyamino acid KLK and a single-stranded oligodeoxynucleotide with alternating sequences of the nucleic acids inosine and cytidine (ODN1a) (4). KLK enables simultaneous uptake of H1 and ODN1a in antigen-presenting cells and provides a platform for hyperefficient Toll-like receptor 9 (TLR-9) ligand recognition of ODN1a. Upon triggering of TLR-9, conventional dendritic cells and monocytes activate the MyD88-NF-κB dependent signaling cascade (5). This leads to secretion of proinflammatory cytokines and finally induction of adaptive immune responses. In addition to the MyD88-NF-κB-dependent pathway, plasmacytoid dendritic cells (pDC) have the unique ability to signal in a MyD88-interferon regulatory factor (IRF7)-dependent way. This yields production of abundant quantities of type I interferon (IFN) (6, 7).
In HIV-infected individuals, vaccine take and durability of the immunity are negatively affected upon manifestation of progressed HIV disease (8). The HIV is detected by the innate immune system, primarily by recognition of viral nucleic acids. Endosomal TLR-7 and -8 and cytosolic RIG-1-like receptors sense single-stranded viral RNA (ssRNA), which leads to secretion of proinflammatory cytokines and type I IFNs (9). Use of a TLR-9 agonist as an adjuvant needs to be tested and analyzed carefully in HIV-infected individuals, since the downstream signaling cascades of TLR-7, TLR-8, and TLR-9 are shared (6, 7).
We conducted a phase II, double-blind, randomized, placebo-controlled trial to evaluate the safety and immunogenicity of H1/IC31 in BCG-vaccinated, HIV-infected adults with a CD4+ lymphocyte count of >350 cells/mm3 (10). The principal finding was that the vaccine was safe and immunogenic. CD4+ lymphocyte counts and viral loads remained stable during the entire study period. The aim of this study was to analyze mRNA expression levels before and shortly after H1/IC31 vaccination and correlate these with vaccine-specific central (TCM) and effector memory CD4+ T cell (TEM) responses. A whole venous blood stimulation assay (10) was used to monitor the vaccine-induced cellular immune response over a follow-up period of 182 days. With use of AmpliSeq and next-generation sequencing the expression changes of 1,388 genes in peripheral blood were determined (11). We describe for the first time that higher expression levels of genes involved in antiviral innate immunity at baseline reduce long-term maintenance of vaccine responses in HIV-infected volunteers.
MATERIALS AND METHODS
Ethics statement.
This study was conducted according to good clinical practice guidelines and the Declaration of Helsinki. Informed consent was obtained from all participants. Detailed descriptions of the ethical approvals can be obtained from Reither et al. (10). The clinical trial was registered in the Pan African Clinical Trials Registry (PACTR) with the identifier PACTR201105000289276.
Participant enrollment and blood collection.
A total of 24 HIV-positive, BCG-vaccinated volunteers with CD4+ T cell counts of >350 cells/mm3 from Bagamoyo, United Republic of Tanzania, were enrolled. Participants were randomly allocated in a ratio of 5:1 to receive placebo or H1/IC31 vaccine. One H1/IC31 vaccine recipient dropped out before the booster vaccination due to pregnancy. Injections of 0.5 ml containing 50 μg Ag85B-ESAT-6 and 500 nmol KLK plus 20 nmol ODN1a (adjuvant) or 0.5 ml Tris buffer (placebo) were administered intramuscularly on study days 0 and 56. For transcriptomics analysis, 2.5-ml samples of peripheral blood were collected via sterile venipuncture in RNA PaxGene Vacutainer tubes (PreAnalytiX) on days 0, 3, and 59. Samples of 9 ml of peripheral blood were collected in sodium heparin tubes and directly processed in whole-blood stimulation assays on days 0, 14, 56, 70, and 182. An extended description of the study design can be obtained from Reither et al. (10).
ICS and analysis by flow cytometry.
The intercellular cytokine staining (ICS) and flow cytometry analyses were described in Reither et al. (10). The present project focused on the analysis of cytokine-producing memory CD4+ T cell subsets. For whole-blood stimulation and ICS, the protocol of Hanekom et al. was followed (12). Briefly, ex vivo whole blood was directly stimulated with either the H1 fusion protein (5 μg/ml; Statens Serum Institute [SSI]) or phytohemagglutinin (5 μg/ml; BioWeb) or left unstimulated. The costimulatory antibodies anti-CD28 and anti-CD49d (0.5 μg/ml; Becton Dickinson [BD] Biosciences) were included in all assay conditions. Whole-blood samples were incubated for 7 h at 37°C and 5% CO2 in an incubator and then treated with brefeldin A (10 μg/ml; Sigma-Aldrich). Subsequently, the whole-blood samples were transferred to a water bath at 37°C and incubated for an additional 5 h. Afterward, the water bath was switched off, and the water and contents were allowed to slowly reach room temperature. At 10 h after the water bath was switched off, the samples were harvested, EDTA (1.8 mM; Sigma-Aldrich) was added, and red blood cells were lysed in fluorescence-activated cell sorter (FACS) lysing solution (BD Biosciences). The remaining white blood cells were preserved in cryo solution containing 50% RPMI 1640 (Lonza), 40% fetal calf serum (BioWest), and 10% dimethyl sulfoxide (DMSO) (Sigma-Aldrich).
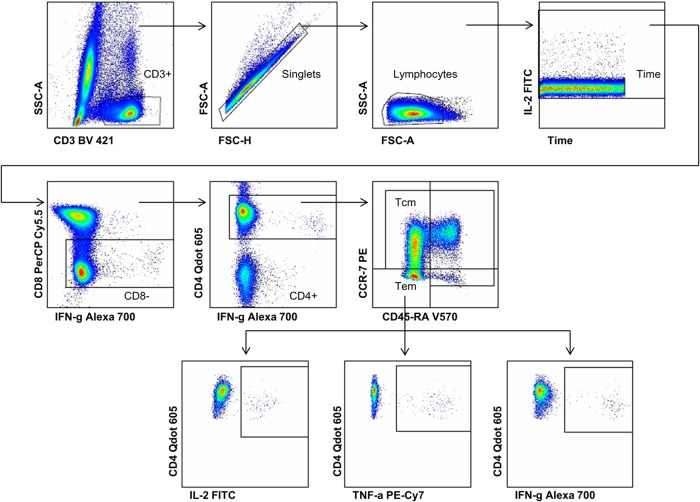
For ICS, samples from each phlebotomy date of one particular participant were thawed simultaneously in a water bath at 37°C. Cells were transferred to tubes containing phosphate-buffered saline (PBS) (BioWhittaker), washed, and permeabilized using Perm/Wash solution (BD Biosciences). The flow cytometry staining was completed with the following anti-human antibody panel: CD3-BV421 (MOPC-21; BD Biosciences), CD4-QDot 605 (S3.5; Invitrogen), CD8-peridinin chlorophyll protein (PerCP)-Cy5.5 (SK1; BD Biosciences), CCR7-phycoerythrin (PE) (150503; eBioscience), IFN-γ-Alexa 700 (B27; BD Biosciences), tumor necrosis factor alpha (TNF-α)-Cy7-PE (Mb11; eBioscience), interleukin-2 (IL-2)-fluorescein isothiocyanate (FITC) (5344.111; BD Biosciences), IL-17-Alexa 647 (SCPL1362; eBioscience), and CD45RA-BV570 (HI100; eBioscience) (see Table 2.1 in the MyFlowCyt Reporting Standard in the supplemental material). For each fluorochrome, a single-stained mouse κ CompBead (BD Biosciences) control was included. The samples were acquired on an LSR II flow cytometer (BD Biosciences, San Jose CA) equipped with green (532 nm), red (640 nm), violet (405 nm), and blue lasers (488 nm) (see Table 3.1 in the MyFlowCyt Reporting Standard in the supplemental material). On average, 900,000 events per sample were recorded. Data analysis was performed using FlowJo software version 10.0.7 (Tree Star). The following gating strategy was used (Fig. 1): CD3-positive cells were gated by plotting CD3-BV421 versus side scatter area (SSC-A). The singlets were selected by plotting forward scatter area (FSC-A) versus forward scatter height (FSC-H), and the lymphocytes were gated with FSC-A versus SSC-A. Time versus IL-2-FITC was gated to exclude bubbles and various fluidics pressures. The IFN-γ-Alexa 700 versus CD8-PerCp-Cy5.5 plot was used to gate the CD8-negative cells. The CD4-positive cells were selected by plotting IFN-γ-Alexa 700 versus CD4-QDot 605. The memory subsets were defined using the CD45RA-BV570 versus CCR7-PE plot. To identify cytokine-positive cells from all 4 memory subsets, each cytokine was plotted against CD4-QDot 605. FlowJo version 10.0.7 was used for the analysis. This project focused on the detection of the cytokines IL-2, IFN-γ, and TNF-α. The production of IL-17 by CD4+ T cells was not significantly different after booster vaccination (10).
FIG 1.
Gating strategy used for flow cytometric analysis of H1-induced memory CD4+ T cell cytokine expression. The plots show the sequential gating hierarchy of one representative sample: CD3+ T cells; single cells; lymphocytes; time; and CD8+ T cells, which were further gated for CD4+ cells and split into all memory subsets on the basis of CCR7 and CD45RA surface markers. The bottom row comprises the plots with the cytokine expression gates for H1 fusion protein-stimulated TEM at day 182. IL-2, TNF-α, and IFN-γ expression levels were measured. TCM, central memory CD4+ T cells; TEM, effector memory CD4+ T cells; H1, hybrid 1.
A detailed description following the MIFlowCyt Reporting Standard for flow cytometry experiments (13) can be accessed in the supplemental material.
Extraction of total RNA.
Total RNA was directly extracted from PaxGene blood RNA tubes using the PaxGene blood RNA kit (PreAnalytiX; Hombrechtikon, Switzerland) according to the manufacturer's protocol. The concentration of total RNA and the RNA integrity number were determined by an Agilent RNA 6000 nano kit (Agilent Technologies, Waldbronn, Germany). RNA samples were stored at −80°C.
AmpliSeq panels.
We used two custom-made AmpliSeq primer panels to quantify gene expression. The Pathway Reporter Panel covered 917 genes, providing a general snapshot of the whole human transcriptome. The rationale and design of this panel are described elsewhere (35). The second AmpliSeq primer panel, the Immune Response Panel, was developed based on a literature-driven collection of 826 genes implicated in innate and adaptive immune responses. Functional enrichment analysis with the biological process (BP) terms of Gene Ontology suggests that the prioritized genes are highly enriched in innate and adaptive immune response pathways. The selected genes were submitted via a web interface for primer design and synthesis using proprietary algorithms (Ampliseq; Life Technologies).
Amplicon-based transcript quantification by semiconductor sequencing.
The amplicon-based transcript quantification was performed using an ion proton semiconductor sequencer. This methodology was previously described by Zhang et al. (11) and strictly followed. Briefly, AmpliSeq libraries were prepared using 30 ng total RNA according to the protocol supplied with the ion AmpliSeq RNA library kit (catalog no. 4472335; Life Technologies, Carlsbad, CA, USA). The amplified and purified libraries were stored at −20°C. Library size distribution and concentration were measured using an Agilent high-sensitivity DNA kit (Agilent Technologies) according to the manufacturer's recommendation. Following library preparation, barcoded samples were pooled and processed together. The multiplexed library (total 8 pM) was linked to ion sphere particles and clonally amplified by emulsion PCR using the Ion PI template OT2 200 kit v3 with the Ion OneTouch 2 instrument according to the manufacturer's protocol (Life Technologies). Sequencing was performed using the Ion PI sequencing 200 kit v3 and the Ion Proton chip I, following the manufacturer's instructions, on the Ion Proton sequencer (Life Technologies). The generated reads were aligned to the Homo sapiens RNA canonical transcript reference hg19 and mapped to the genes of the corresponding AmpliSeq panel using the Torrent Mapping Alignment Program. Simultaneously, single nucleotide polymorphisms (SNPs) within the amplicons were identified during this process using the Ion Torrent Variant Caller.
Statistical analysis.
(i) Analysis of memory CD4+ T cells.
A two-sided Wilcoxon signed-rank test was performed to test for the significance of the polyfunctional memory CD4+ T cell subset between study days. The participants' responses to the vaccine were calculated using the Mimosa package within R software (14, 15). After correction for the T cell response obtained from unstimulated controls, absolute counts of cytokine (IFN-γ and/or IL-2 and/or TNF-α)-expressing TCM or TEM and counts of TCM or TEM negative for cytokine expression were compared between day 0 and each follow-up (day 14, day 56, day 70, and day 182) (see Table 2). Final responders were defined based on a comparison of day 0 to day 182 and with a false discovery rate (FDR) of >0.0001 for a nonresponder, an FDR of >1015 for an intermediate responder, and an FDR of <1015 for a high responder.
TABLE 2.
Overview of responder groups based on either TCM or TEM
| T cell type | No. of nonresponders/intermediate responders/high responders at: |
|||
|---|---|---|---|---|
| Day 14 | Day 56 | Day 70 | Day 182 | |
| TCM | 6/6/7 | 11/3/5 | 5/6/8 | 8/5/6 |
| TEM | 3/5/11 | 4/5/10 | 2/2/5 | 4/6/9 |
(ii) AmpliSeq data analysis.
AmpliSeq data were analyzed based on negative binomial distribution using the Bioconductor edgeR package (16). For each vaccine responder group, we performed differential expression analysis through the time course, comparing the two postvaccination time points to the prevaccination time point. Furthermore, gene expression was compared between the vaccine responder groups in order to identify values that differed from baseline. Standard settings were used, and only those genes with expression higher than 5 counts per million reads in at least 3 samples and an FDR of <0.1 were considered differentially expressed. Annotation of differentially expressed genes was performed using the previously published blood transcription modules (BTMs) (17). Statistical analysis was performed and plots were made using R version 3.1.0 (15).
RESULTS
H1/IC31 phase II clinical trial design.
We conducted a phase II, double-blind, randomized, placebo-controlled trial in Bagamoyo, Tanzania (10). Twenty-four HIV-positive volunteers were enrolled and vaccinated twice with H1/IC31 (n = 20) or placebo (n = 4) at baseline (day 0) and day 56. One H1/IC31-vaccinated volunteer dropped out due to pregnancy. Whole-blood samples for transcriptome analysis were taken on study days 0, 3, and 59 after each vaccination. Whole-blood samples for immunogenicity assays were taken on study days 0, 14, 56, 70, and 182 for long-term follow-up. All volunteers were BCG vaccinated prior to the study and were antiretroviral therapy (ART) naive with CD4 counts of >350 cells/mm3 (Table 1). Generally, the H1/IC31 vaccine was safe and well tolerated with no significant impact on the CD4+ lymphocyte count and HIV load after vaccination.
TABLE 1.
H1/IC31 clinical trial overview and results
| Characteristica | Value for: |
|
|---|---|---|
| Placebo group (n = 4) | H1/IC31 vaccinated group (n = 19) | |
| Age (median [IQR]) (yr) | 38 (28−49) | 39 (34−45) |
| Gender (F/M) | 3/1 | 9/10 |
| No. (%) with prior BCG vaccination (self-report) | 4 (100) | 19 (100) |
| No. (%) ART naive | 4 (100) | 19 (100) |
| CD4 count (median [IQR]) from baseline (cells/mm3) | 575.3 (425–617)b | 656.9 (532−745)b |
| Viral load (median [IQR]) from baseline (copies/ml) | 33,802 (514–49,875)b | 37,932 (8,754−50,950)b |
IQR, interquartile range; F, female; M, male.
No significant difference over the 182-day follow-up.
Cytokine-producing memory CD4+ T cell subsets.
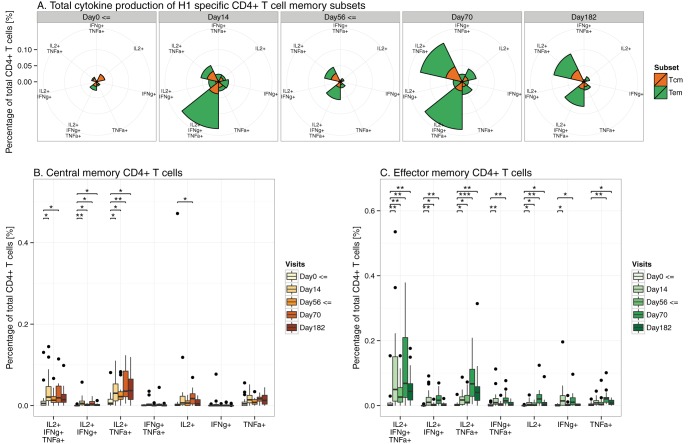
Based on chemokine receptor CCR7 and surface marker CD45RA expression, two subsets of CD4+ memory T cells, TCM and TEM, were identified (Fig. 1) (18). We measured the development and magnitude of the H1/IC31-induced memory CD4+ T cell subsets expressing IL-2, IFN-γ, and TNF-α following in vitro stimulation with the H1 protein. At day 182, TCM and TEM were significantly elevated compared to levels in placebo controls (see Fig. S1 in the supplemental material). To measure the quality of the immune response, we discriminated the memory CD4+ T cell subsets according to their polyfunctional cytokine expression profile. Figure 2A displays the mean percentages of cytokine expression of all memory subsets according to their polyfunctionality. In agreement with Reither et al. (10), the H1/IC31-specific TCM and TEM were predominantly trifunctional or bifunctional, expressing IL-2 and TNF-α.
FIG 2.
Cytokine expression profile of memory CD4+ T cell subsets following H1 fusion protein stimulation. Volunteers receiving H1/IC31 at days 0 and 56 are included. (A) Mean percentages of all memory CD4+ T cell subsets on the basis of all possible combinations of IL-2, IFN-γ, and TNF-α expression are given. Radii and not area of each segment represent the percentages of CD4+ T cells. TCM (B) and TEM (C) expressing all combinations of IL-2, IFN-γ, and TNF-α expression following stimulation with H1. P values correspond to significance testing comparing each study day to study day 0 (*, P < 0.0125; **, P < 0.001; ***, P < 0.0001). A two-sided Wilcoxon signed-rank test with the Bonferroni correction was applied. TCM, central memory CD4+ T cells; TEM, effector memory CD4+ T cells; H1, hybrid 1; ⇐, vaccination days.
In Fig. 2B and C, TCM and TEM are displayed separately. In a comparison of day 0 to day 182, statistically significant increases of bifunctional TCM expressing IL-2 with TNF-α (P < 0.05) were observed. This was also observed in bifunctional TCM expressing IL-2 with IFN-γ, although at extremely low percentages.
Trifunctional and bifunctional (IL-2 and TNF-α) TEM were most significantly elevated in the comparison of day 182 to day 0 (P < 0.01), followed by monofunctional (IL-2 or TNF-α) TEM (P < 0.05).
Definition of vaccine responder groups.
To investigate potential innate immune mechanisms that yield differential vaccine immunogenicity in our study participants, we divided the volunteers according to their cytokine responses (IFN-γ, IL-2, and/or TNF-α), comparing H1-specific TCM and TEM.
An overview of potential responder groupings comparing each study day to the baseline at day 0 is given in Table 2. In a comparison of day 70 to day 182, several volunteers shifted from responders to nonresponders, indicating discriminative memory maintenance.
No significant difference in relation to the viral load and CD4+ lymphocyte counts between these groups was observed (Table 3).
TABLE 3.
Demographics including information about HIV infection status of final responder groups based on the comparison of day 0 to day 182
| Day 182 responder group | No. of volunteers | Age (yr)a | No. F/no. Mb | Viral load (log2)a | CD4+ count (cells/mm3)a |
|---|---|---|---|---|---|
| Placebo | 4 | 38 ± 13 | 3/1 | 11.5 ± 5.3 | 575 ± 250 |
| TCM | |||||
| High responder | 6 | 35 ± 5 | 2/4 | 12.1 ± 3.8 | 823 ± 266 |
| Intermediate responder | 5 | 46 ± 5 | 2/3 | 14.0 ± 3.1 | 529 ± 64 |
| Nonresponder | 8 | 38 ± 6 | 6/2 | 14.3 ± 2.7 | 618 ± 181 |
| TEM | |||||
| High responder | 9 | 38 ± 7 | 4/5 | 12.3 ± 3.4 | 698 ± 284 |
| Intermediate responder | 6 | 40 ± 9 | 2/4 | 14.4 ± 3.4 | 604 ± 90 |
| Nonresponder | 4 | 39 ± 5 | 4/0 | 14.9 ± 1.2 | 644 ± 226 |
Data are means ± SD.
F, female; M, male.
Grouping the volunteers based on a comparison of day 0 to day 182 allowed us to identify the innate immune mechanisms that potentially lead to improved TCM and TEM maintenance.
Gene expression data.
On average, 4,686,802 reads (95% confidence interval [CI], ±712,607 reads) per sample were generated and mapped to either the 826 genes of the AmpliSeq Immune Response Panel or the 917 genes of the Pathway Reporter Panel. A dynamic range of 5 orders of magnitude was observed, ranging from genes with no detectable expression like IFN-κ to genes expressing more than 160,000 reads like lysozyme and CD74. Both AmpliSeq panels shared 355 genes, whose transcript abundance measurements closely correlated (R2 = 0.9829).
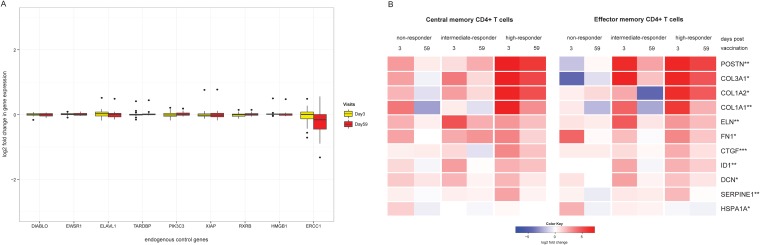
The expression levels of nine endogenous control genes did not differ significantly between the 24 volunteers included here across all time points, supporting the robustness of the AmpliSeq-based targeted transcriptome measurement approach (Fig. 3A).
FIG 3.
(A) Gene expression of endogenous control genes. Variations in expression levels of nine control genes between subjects and over time and log2-fold changes between baseline and the two visits after each vaccination for all 24 volunteers are shown. (B) Heat map showing vaccine-induced differential gene expression levels between TCM and TEM responder groups. Log2-fold changes in gene expression levels 3 or 59 days after vaccination compared to prevaccination levels are shown. Differentially expressed genes were identified using edgeR with an FDR of <0.1 (**, significant for TCM and TEM; *, significant for TEM only). Only changes in gene expression 3 days after vaccination were significant.
Vaccine-induced differential gene expression.
Next we addressed if early changes in the whole-blood gene expression levels of the targeted 1,388 genes might distinguish the three responder groups of each memory subset. The results are shown in Fig. 3B, and more detailed information is provided in Table S1 in the supplemental material. TCM and TEM intermediate and high responders showed upregulated gene expression levels 3 days postvaccination. For TCM responders, 5 genes, namely, COL1A1, ELN, CTGF, SERPINE1, and POSTN, were identified as upregulated. TEM responders had an additional 5 genes, including COL3A1, COL1A2, FN1, ID1, and DCN, with increased expression levels.
Most of the 10 differentially expressed genes are either annotated in BTMs involved in integrin interactions or extracellular matrix and cell adhesion (COL1A1, COL1A2, COL3A1, CTGF, FN1, and POSTN) or known to be part of extracellular matrix remodeling (ELN or SERPINE1). ID1 is involved in leukocyte differentiation. HSPA1A is the only gene which was upregulated in a TEM nonresponder. This gene encodes a heat shock protein and is involved in the stress response. No differential gene expression was observed in the placebo controls.
Differential immune activation at baseline.
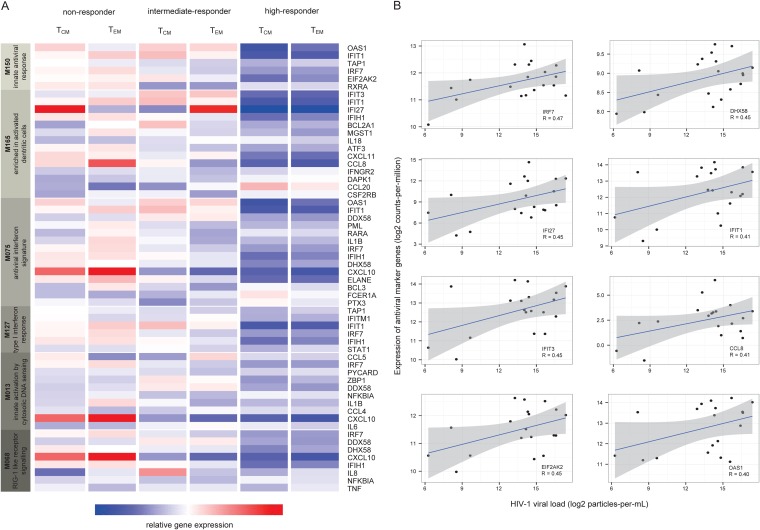
Next we addressed if gene expression levels at baseline before the first vaccination could distinguish the responder groups. Table 4 shows an overview of all genes that differed significantly at baseline between the vaccine responder groups based either on TCM or TEM. Compared to levels in nonresponders, higher gene expression levels were observed for ITGB4, CLEC1C, and CXCL6 for TCM high responders and for PRKDC, CD19, DNAJB5, MARCO, and IRS2 for TEM high responders. TGM2, OAS1, and IFI27 showed the lowest expression levels among TCM high responders, whereas AICDA, NFATC2, IFI27, IFIT3, IFIT1, and CXCL10 were expressed at lower levels in TEM high responders. Tissue transglutaminase (TGM) has a role in HIV pathogenesis and enhanced levels of its degradation product, ε(γ-glutamyl)lysine, are found in the blood of patients with progressed HIV disease (19). AICDA and NFATC2 take part in the antiviral immune response with the roles of affinity maturation and class switching of antibodies and inhibition of viral replication (19, 20). According to annotation with BTMs, the lower expressed genes indicated in bold above are involved in innate antiviral immunity. We analyzed the expression levels of all genes available in both AmpliSeq panels that belonged to these BTMs (Fig. 4A). Clearly, at baseline both nonresponder groups showed higher transcription levels of members of BTM M150 (innate antiviral response), M165 (enriched in activated dendritic cells), M75 (antiviral IFN signature), M127 (type I interferon response), M13 (innate activation by cytosolic DNA sensing), and M68 (RIG-1-like receptor signaling).
TABLE 4.
Genes which are differentially expressed between the different vaccine responder groups for TCM and TEM
| T cell type | Genes with expression levels:a |
|
|---|---|---|
| Higher than those in nonresponders | Lower than those in nonrespondersb | |
| TCM | ITGB4 | TGM2 |
| CLEC1A | OAS1 | |
| CXCL6 | IFI27 | |
| TEM | PRKDC | AICDA |
| CD19 | NFATC2 | |
| DNAJB5 | IFI27 | |
| MARCO | IFIT3 | |
| IRS2 | IFIT1 | |
| CXCL10 | ||
Differentially expressed genes were identified using edgeR with an FDR of <0.1 and are grouped as having higher or lower expression levels in the high-responder group than in the nonresponder group.
Genes in bold are involved in innate antiviral immunity.
FIG 4.
Differential gene expression at baseline prior to vaccination. (A) Innate antiviral signature at baseline. The expression levels of the five genes involved in innate antiviral immunity with their corresponding members of BTMs are shown for TCM and TEM responder groups. (B) Correlation of innate antiviral markers and viral load. Pearson correlation coefficients and 95% confidence intervals are shown. Only genes with Pearson correlation coefficients of >0.4 were considered. TCM, central memory CD4+ T cells; TEM, effector memory CD4+ T cells.
The expression levels of genes involved in innate immunity to viruses (IRF7, IFI27, IFIT3, EIF2AK2, DHX58, IFIT1, CCL8, and OAS1) showed an association with HIV-1 viral loads (Fig. 4B).
Toll-like receptor 8 variant.
The AmpliSeq approach allows for nucleotide sequencing of the amplified fragments using the Ion Torrent approach. We are aware that this study was not designed (e.g., powered) to assess the association of polymorphic genes with vaccine-induced immune responses. One single nucleotide polymorphism (SNP) in TLR-8 (rs3764880) was previously reported to result in slower disease progression in HIV-infected individuals and reduced activation of the NF-κB pathway (21). The functional variant of TLR-8 showed the A1G polymorphism (rs3764880) that alters the start ATG codon of TLR-8 into a GTG triplet. The resulting truncated TLR-8 (1,038 amino acids [aa]) exhibits a shorter signal peptide. Seven volunteers expressed the TLR-8 A1G; of these, three volunteers harbored the homozygous TLR-8 A1G. Interestingly, none of the volunteers with the TLR-8 A1G belonged to the TEM nonresponder group, and all three subjects expressing the homozygous TLR-8 A1G were TEM high responders. Importantly, subjects with the homozygous TLR-8 A1G have a reduced expression of antiviral gene signatures at baseline (Fig. 5A). Independent of the responder grouping, at day 182 volunteers with the homozygous TLR-8 A1G have on average the highest percentages of trifunctional and bifunctional (IL-2 and TNF-α) TCM and TEM (Fig. 5B and C).
FIG 5.
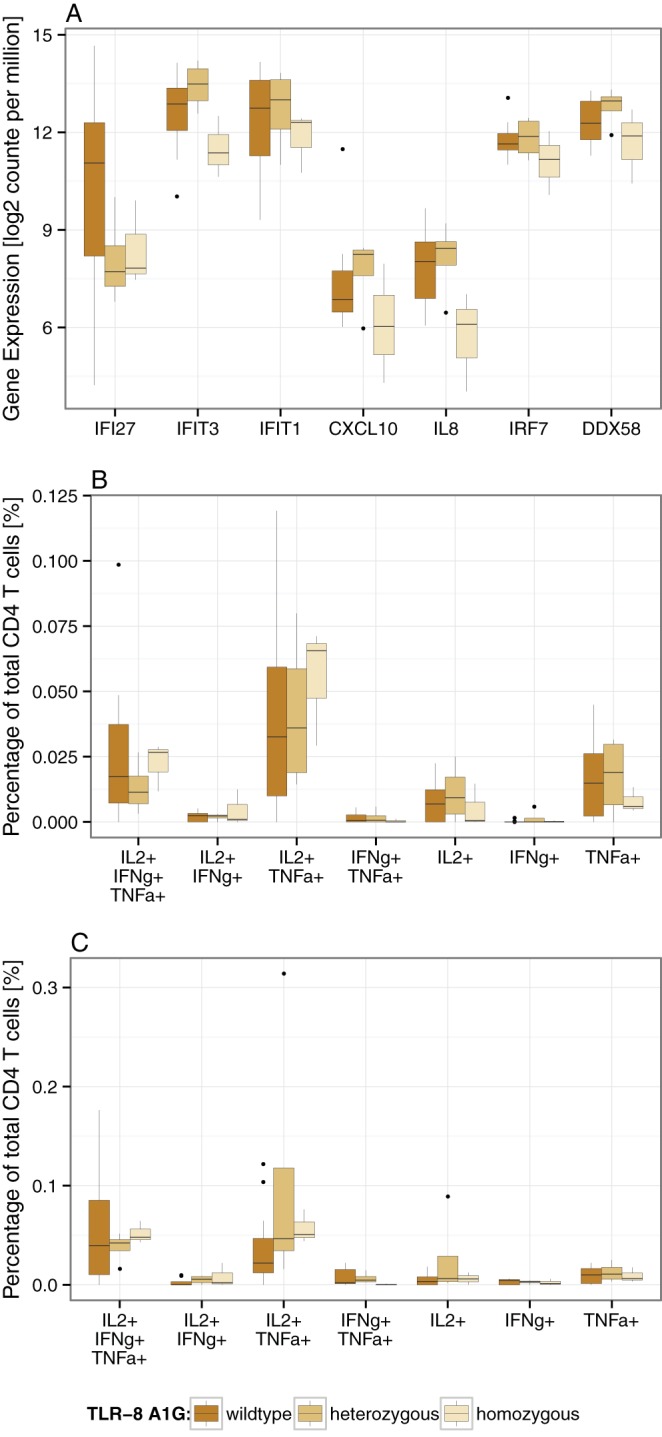
(A) Baseline expression of genes implicated in innate antiviral immunity comparing volunteers with either wild-type, heterozygous, or homozygous TLR-8 A1G. (B and C) Cytokine expression profiles of TCM and TEM following H1 fusion protein stimulation at day 182. The differences between volunteers with wild-type, heterozygous, and homozygous TLR-8 A1G are presented. Due to low numbers of volunteers with homozygous TLR-8 A1G, no statistical test was applied.
DISCUSSION
The study presented here provides the first insights into the possible link between chronic immune activation in HIV infection and lack of maintenance of vaccine-induced TCM and TEM responses. Data on the impact of chronic, untreated HIV infection on experimental and routine vaccination outcomes are scarce (22). The identification of immune-related genes that are expressed before vaccination or induced shortly after vaccination influencing cellular immune responses in HIV-positive volunteers will be of great value (23).
Determinants of induction and long-term maintenance of memory CD4+ T cell responses generally in humans are only partially known (24). The H1/IC31 vaccine has been tested in several phase I clinical trials, including HIV-negative and BCG-unvaccinated and BCG-vaccinated and M. tuberculosis-exposed European volunteers (4, 25). In the BCG- and M. tuberculosis-naive European volunteers, IFN-γ-producing H1-specific T cells were detectable by ELISpot analysis until 131 weeks after the first vaccination. Only limited retraction of IFN-γ production in cell culture supernatants from H1-stimulated peripheral blood mononuclear cells (PBMCs) was measured between study weeks 14 and 131 (4). In the current study, we observed an expansion of H1-specific TCM and TEM in all volunteers at study day 70, which is 2 weeks after the booster vaccination. However, when followed up until day 182, a retraction of H1-specific TCM and TEM was observed. Interestingly, this retraction differed greatly between volunteers, which allowed us to group them based on the comparison of H1/IC31-induced TCM and TEM responses on day 182 in relation to those on day 0.
Compared with microarray analyses, the AmpliSeq approach has a higher sensitivity and dynamic range, and its results compare precisely with RNA sequencing-derived whole-transcriptome data (11). Results obtained with both gene panels strongly supported the idea that the AmpliSeq-based transcriptome monitoring approach is very robust and results in highly reproducible data sets suitable for high-throughput analysis of clinical samples. Here, to our knowledge, we have employed this targeted transcriptome monitoring tool in an experimental vaccine trial for the first time.
Higher expression levels of genes involved in the extracellular matrix and integrin interactions and cell adhesion were observed in volunteers who maintained H1-specific TCM and TEM responses until day 182. Searching the literature, we found that cells involved in wound healing like alternatively activated macrophages (26), γδ-T cells (27), monocyte-derived multipotential cells (28), fibrocytes (17), and endothelial progenitor cells (29) have all been described to express these particular genes.
Muyanja et al. showed that the cellular and humoral immune responses to the highly efficacious live yellow fever vaccine YF-17D in East African individuals were substantially lower than those of European volunteers treated identically (30). Prior to vaccination, the East African volunteers presented an increased activated immune microenvironment with higher frequencies of exhausted and activated natural killer cells, differentiated B and T cells, and proinflammatory monocytes (30). We therefore compared the gene expression profiles measured at baseline between our responder groups. Significantly higher expression levels of genes involved in innate antiviral immune responses like type I IFN signaling and viral sensing through cytosolic RIG-1-like receptors were observed in the nonresponders. A close correlation between HIV loads and expression levels of this group of genes indicates that the chronic HIV infection is driving the higher expression levels of these antiviral responses. In line with these results, HIV disease progressors are known to display a type I IFN chronic exposure signature when contrasted to HIV disease elite controllers (31, 32). The study by Negishi et al. reported that activation of RIG-1-like receptors resulted in selective suppression of TLR signaling and thus supports our findings (33). The HIV was found to specifically infect activated memory CD4+ T cells with potential consequences for the maintenance of vaccine-specific memory responses (34). In conclusion, higher activation of the components of the innate immunity due to HIV stimulation at baseline results in impaired H1-specific TCM and TEM maintenance.
In addition to access to the expression level, the AmpliSeq approach also provides access to the nucleotide sequence of the respective amplified gene fragment. This enabled us to identify volunteers carrying the previously known TLR-8 A1G single nucleotide sequence variant. Volunteers expressing this TLR-8 A1G gene on both chromosomes had lower gene expression levels of the innate antiviral immune response genes. This was linked to longer maintenance of vaccine-specific TEM responses until day 182 compared to that for the wild-type TLR-8 carriers.
The inverse relationship between chronic innate antiviral immune activation by HIV and sustained H1/IC31-induced vaccine-specific cellular immune responses has high relevance for future vaccine development and monitoring programs. Patients receiving ART potentially present with lower activation of innate antiviral immunity, and thus the effect of treating patients with ART prior to vaccination has to be evaluated. The presence of the TLR-8 A1G variant in HIV-infected individuals might provide an immune genetic background, supporting a more desired vaccination outcome. This observation warrants future studies using larger numbers of HIV-infected and -noninfected volunteers undergoing experimental or routine vaccination.
ACKNOWLEDGMENTS
This work was supported by the European Developing Countries Clinical Trials Partnership (EDCTP) (grant IP.2009.32080.002) and the State Secretariat for Education and Research. The AmpliSeq analysis was funded by the Roche Innovation Center Basel.
We are grateful to all study participants. We also thank the sponsor, the Statens Serum Institut, and the members of the Data Safety Monitoring Board: Anthony Hawkridge, Jaap van Dissel, Clive Gray, Eyasu Makonnen, and Gibson Kibiki. We appreciate the support of Biolytix AG (www.biolytix.ch), who provided their RNA extraction platform.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00092-15.
REFERENCES
- 1.Omer SB, Orenstein WA, Koplan JP. 2013. Go big and go fast—vaccine refusal and disease eradication. N Engl J Med 368:1374–1376. doi: 10.1056/NEJMp1300765. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2013. Global tuberculosis report. http://www.who.int/tb/publications/global_report/en/.
- 3.Daley CL, Small PM, Schecter GF, Schoolnik GK, McAdam RA, Jacobs WR Jr, Hopewell PC. 1992. An outbreak of tuberculosis with accelerated progression among persons infected with the human immunodeficiency virus. N Engl J Med 326:231–235. doi: 10.1056/NEJM199201233260404. [DOI] [PubMed] [Google Scholar]
- 4.van Dissel JT, Soonawala D, Joosten SA, Prins C, Arend SM, Bang P, Tingskov PN, Lingnau K, Nouta J, Hoff ST, Rosenkrands I, Kromann I, Ottenhoff TH, Doherty TM, Andersen P. 2010. Ag85B-ESAT-6 adjuvanted with IC31 promotes strong and long-lived Mycobacterium tuberculosis specific T cell responses in naive human volunteers. Vaccine 28:3571–3581. doi: 10.1016/j.vaccine.2010.02.094. [DOI] [PubMed] [Google Scholar]
- 5.Aichinger MC, Ginzler M, Weghuber J, Zimmermann L, Riedl K, Schütz G, Nagy E, von Gabain A, Schweyen R, Henics T. 2011. Adjuvating the adjuvant: facilitated delivery of an immunomodulatory oligonucleotide toTLR9 by a cationic antimicrobial peptide in dendritic cells. Vaccine 29:426–436. doi: 10.1016/j.vaccine.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Blasius AL, Beutler B. 2010. Intracellular Toll-like receptors. Immunity 32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki T, Kawai T. 2014. Toll-like receptor signaling pathways. Front Immunol 5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okulicz JF, Mesner O, Ganesan A, O'Bryan TA, Deiss RG, Agan BK. 2014. Hepatitis B vaccine responsiveness and clinical outcomes in HIV controllers. PLoS One 9:e105591. doi: 10.1371/journal.pone.0105591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki A. 2012. Innate immune recognition of HIV-1. Immunity 37:389–398. doi: 10.1016/j.immuni.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reither K, Katsoulis L, Beattie T, Gardiner N, Lenz N, Said K, Mfinanga E, Pohl C, Fielding KL, Jeffrey H, Kagina BM, Hughes EJ, Scriba TJ, Hanekom WA, Hoff ST, Bang P, Kromann I, Daubenberger C, Andersen P, Churchyard GJ. 2014. Safety and immunogenicity of H1/IC31, an adjuvanted TB subunit vaccine, in HIV-infected adults with CD4+ lymphocyte counts greater than 350 cells/mm3: a phase II, multi-centre, double-blind, randomized, placebo-controlled trial. PLoS One 9:e114602. doi: 10.1371/journal.pone.0114602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Schindler T, Küng E, Ebeling M, Certa U. 2014. Highly sensitive amplicon-based transcript quantification by semiconductor sequencing. BMC Genomics 15:565. doi: 10.1186/1471-2164-15-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanekom WA, Hughes J, Mavinkurve M, Mendillo M, Watkins M, Gamieldien H, Gelderbloem SJ, Sidibana M, Mansoor N, Davids V, Murray RA, Hawkridge A, Haslett PA, Ress S, Hussey GD, Kaplan G. 2004. Novel application of a whole blood intracellular cytokine detection assay to quantitate specific T-cell frequency in field studies. J Immunol Methods 291:185–195. doi: 10.1016/j.jim.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 13.Lee JA, Spidlen J, Boyce K, Cai J, Crosbie N, Dalphin M, Furlong J, Gasparetto M, Goldberg M, Goralczyk EM, Hyun B, Jansen K, Kollmann T, Kong M, Leif R, McWeeney S, Moloshok TD, Moore W, Nolan G, Nolan J, Nikolich-Zugich J, Parrish D, Purcell B, Qian Y, Selvaraj B, Smith C, Tchuvatkina O, Wertheimer A, Wilkinson P, Wilson C, Wood J, Zigon R, International Society for Advancement of Cytometry Data Standards Task Force, Scheuermann RH, Brinkman RR. 2008. MIFlowCyt: the minimum information about a flow cytometry experiment. Cytometry A 73A:926–930. doi: 10.1002/cyto.a.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finak G, McDavid A, Chattopadhyay P, Dominguez M, De Rosa S, Roederer M, Gottardo R. 2014. Mixture models for single-cell assays with applications to vaccine studies. Biostatistics 15:87−101. doi: 10.1093/biostatistics/kxt024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.R Core Team. 2014. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 16.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. 2006. GenePattern 2.0. Nat Genet 38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 17.Li G, Cheng M, Nunoya J, Cheng L, Guo H, Yu H, Liu YJ, Su L, Zhang L. 2014. Plasmacytoid dendritic cells suppress HIV-1 replication but contribute to HIV-1 induced immunopathogenesis in humanized mice. PLoS Pathog 10:e1004291. doi: 10.1371/journal.ppat.1004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708−712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 19.Farrow MA, Kim EY, Wolinsky SM, Sheehy AM. 2011. NFAT and IRF proteins regulate transcription of the anti-HIV gene, APOBEC3G. J Biol Chem 286:2567–2577. doi: 10.1074/jbc.M110.154377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu W, Santini PA, Sullivan JS, He B, Shan M, Ball SC, Dyer WB, Ketas TJ, Chadburn A, Cohen-Gould L, Knowles DM, Chiu A, Sanders RW, Chen K, Cerutti A. 2009. HIV-1 evades virus-specific IgG2 and IgA class switching by targeting systemic and intestinal B cells via long-range intercellular conduits. Nat Immunol 10:1008–1017. doi: 10.1038/ni.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh DY, Taube S, Hamouda O, Kücherer C, Poggensee G, Jessen H, Eckert JK, Neumann K, Storek A, Pouliot M, Borgeat P, Oh N, Schreier E, Pruss A, Hattermann K, Schumann RR. 2008. A functional Toll-like receptor 8 variant is associated with HIV disease restriction. J Infect Dis 198:701–709. doi: 10.1086/590431. [DOI] [PubMed] [Google Scholar]
- 22.Kernéis S, Launay O, Turbelin C, Batteux F, Hanslik T, Boëlle PY. 2014. Long-term immune responses to vaccination in HIV-infected patients: a systematic review and meta-analysis. Clin Infect Dis 58:1130–1139. doi: 10.1093/cid/cit937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seder RA, Hill AV. 2000. Vaccines against intracellular infections requiring cellular immunity. Nature 406:793–798. doi: 10.1038/35021239. [DOI] [PubMed] [Google Scholar]
- 24.Farber DL, Yudanin NA, Restifo NP. 2014. Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol 14:24–35. doi: 10.1038/nri3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Dissel JT, Soonawala D, Joosten SA, Prins C, Arend SM, Bang P, Tingskov PN, Lingnau K, Nouta J, Hoff ST, Rosenkrands I, Kromann I, Ottenhoff TH, Doherty TM, Andersen P. 2011. Ag85B-ESAT-6 adjuvanted with IC31 promotes strong and long-lived Mycobacterium tuberculosis specific T cell responses in volunteers with previous BCG vaccination or tuberculosis infection. Vaccine 29:2100–2109. doi: 10.1016/j.vaccine.2010.12.135. [DOI] [PubMed] [Google Scholar]
- 26.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. 2013. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol 229:176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 27.Rani M, Zhang Q, Schwacha MG. 2014. Gamma delta T cells regulate wound myeloid CELL activity after burn. Shock 42:133–141. doi: 10.1097/SHK.0000000000000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seta N, Kuwana M. 2010. Derivation of multipotent progenitors from human circulating CD14+ monocytes. Exp Hematol 38:557–563. doi: 10.1016/j.exphem.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Gremmels H, Fledderus JO, van Balkom BW, Verhaar MC. 2011. Transcriptome analysis in endothelial progenitor cell biology. Antioxid Redox Signal 15:1092–1042. doi: 10.1089/ars.2010.3594. [DOI] [PubMed] [Google Scholar]
- 30.Muyanja E, Ssemaganda A, Ngauv P, Cubas R, Perrin H, Srinivasan D, Canderan G, Lawson B, Kopycinski J, Graham AS, Rowe DK, Smith MJ, Isern S, Michael S, Silvestri G, Vanderford TH, Castro E, Pantaleo G, Singer J, Gillmour J, Kiwanuka N, Nanvubya A, Schmidt C, Birungi J, Cox J, Haddad EK, Kaleebu P, Fast P, Sekaly RP, Trautmann L, Gaucher D. 2014. Immune activation alters cellular and humoral responses to yellow fever 17D vaccine. J Clin Invest 124:3147–3158. doi: 10.1172/JCI75429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hyrcza MD, Kovacs C, Loutfy M, Halpenny R, Heisler L, Yang S, Wilkins O, Ostrowski M, Der SD. 2007. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J Virol 81:3477–3486. doi: 10.1128/JVI.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rotger M, Dalmau J, Rauch A, McLaren P, Bosinger SE, Martinez R, Sandler NG, Roque A, Liebner J, Battegay M, Bernasconi E, Descombes P, Erkizia I, Fellay J, Hirschel B, Miró JM, Palou E, Hoffmann M, Massanella M, Blanco J, Woods M, Günthard HF, de Bakker P, Douek DC, Silvestri G, Martinez-Picado J, Telenti A. 2011. Comparative transcriptomics of extreme phenotypes of human HIV-1 infection and SIV infection in sooty mangabey and rhesus macaque. J Clin Invest 121:2391–2400. doi: 10.1172/JCI45235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Negishi H, Yanai H, Nakajima A, Koshiba R, Atarashi K, Matsuda A, Matsuki K, Miki S, Doi T, Aderem A, Nishio J, Smale ST, Honda K, Taniguchi T. 2012. Cross-interference of RLR and TLR signaling pathways modulates antibacterial T cell responses. Nat Immunol 13:659–666. doi: 10.1038/ni.2307. [DOI] [PubMed] [Google Scholar]
- 34.Okoye AA, Picker LJ. 2013. CD4+ T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol Rev 254:54–64. doi: 10.1111/imr.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang JD, Küng E, Boess F, Certa U, Ebeling M. 2015. Pathway reporter genes define molecular phenotypes of human cells. BMC Genomics 16:342. doi: 10.1186/s12864-015-1532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]