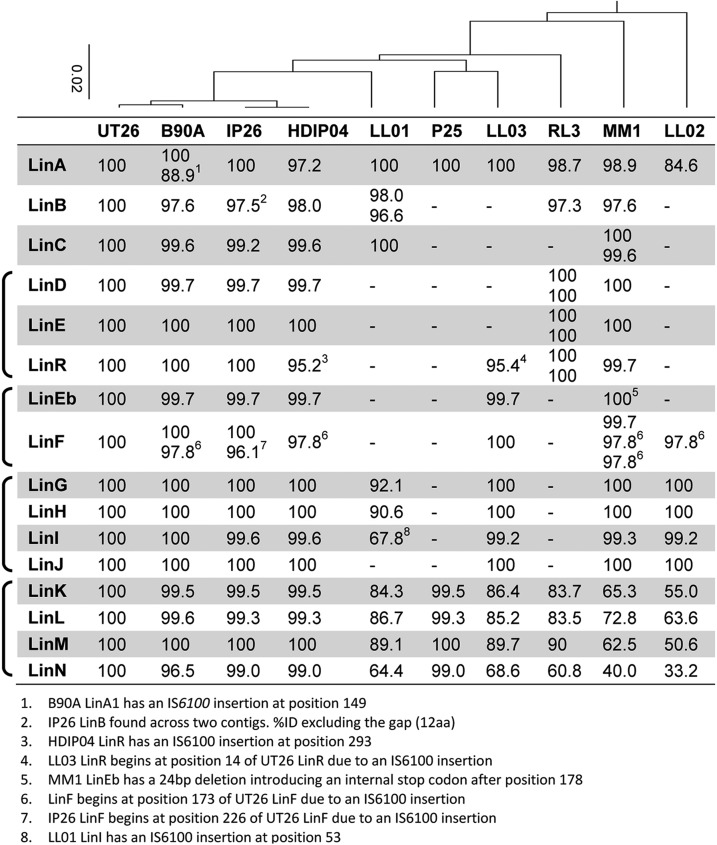
Figure 3.
lin pathway composition of studied strains. Presence of lin genes in the 10 HCH-degrading strains studied, along with the percent identity of each encoded protein to the equivalent in UT26. Genes grouped in operons are indicated with braces to the left of the gene names. Multiple values indicate duplicate copies of the gene in a strain. The neighbor-joining phylogeny above the strain names was produced from a clustalw alignment of the 16S rDNA sequences (UT26: AF039168, B90A: AY519129, IP26: EF190507, HDIP04: EF424393, LL01: JN646865, P25: EU781657, LL03: JN695620, RL-3: EF207155, MM-1: CP004036;G432_r19183, LL02: JN695619) of each strain with E. coli (ECK3843) as an outgroup (not shown). HCH, hexachlorocyclohexane.