Figure 2.
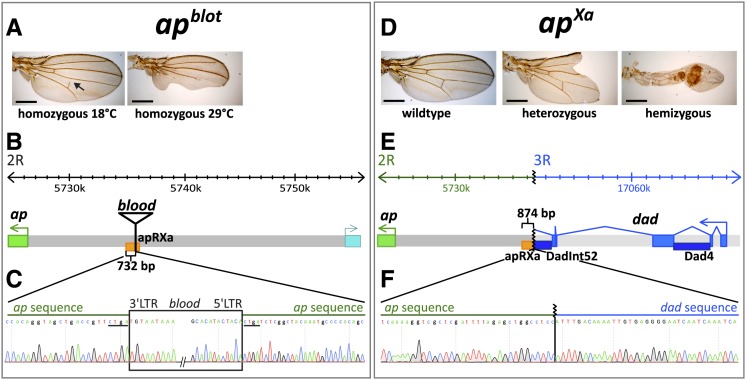
The mutations apblot and apXa affect the ap wing enhancer region. (A) Temperature-sensitive wing phenotypes obtained for the homozygous apblot allele. At 18°, less than 30% of the wings are affected and most of them only show a disruption of the posterior crossvein (arrow). At 29°, ∼70% of the wings have a phenotype. In many of them, the posterior compartment is severely affected. (B) At the top of the panel, the coordinates of the apterous locus are indicated. The insertion site of blood, a retrotransposable element, within the apRXa wing enhancer is depicted. (C) Sequence data close to the insertion site of the blood element in apblot. The insertion causes a four bp duplication (CTGA, underlined). Exact coordinates of the 4 bp duplication: 2R:5735176.0.5735179 (Flybase Release FB2014_06). (D) Preparations of wild type and apXa mutant wings. All apXa/+ flies show a dominant phenotype: the distal part of the wing blade is lost and the characteristic mitten phenotype is formed. In hemizygous condition, the wing tissue of apXa/apDG3 flies forms a short tube-like structure. Margin bristles are absent except for sometimes a few at the tip. All scale bars are 50 µm. (E) Molecular characteristics of the apXa mutation. A reciprocal translocation involving the right arms of the second and third chromosome causes a breakpoint just upstream of the apRXa wing enhancer (indicated in orange) and juxtaposes the daughters against dpp (dad) locus (indicated in blue) next to the ap gene. The dark blue rectangles represent the well-studied cis-regulatory elements Dadint52 and Dad4 which are active in the wing disc (Weiss et al. 2010). (F) Chromatograph of the apXa sequence across the rearrangement break point. The coordinates of the breakpoints are: 2R:5375319 and 3R:17065902 (Flybase Release FB2014_06).