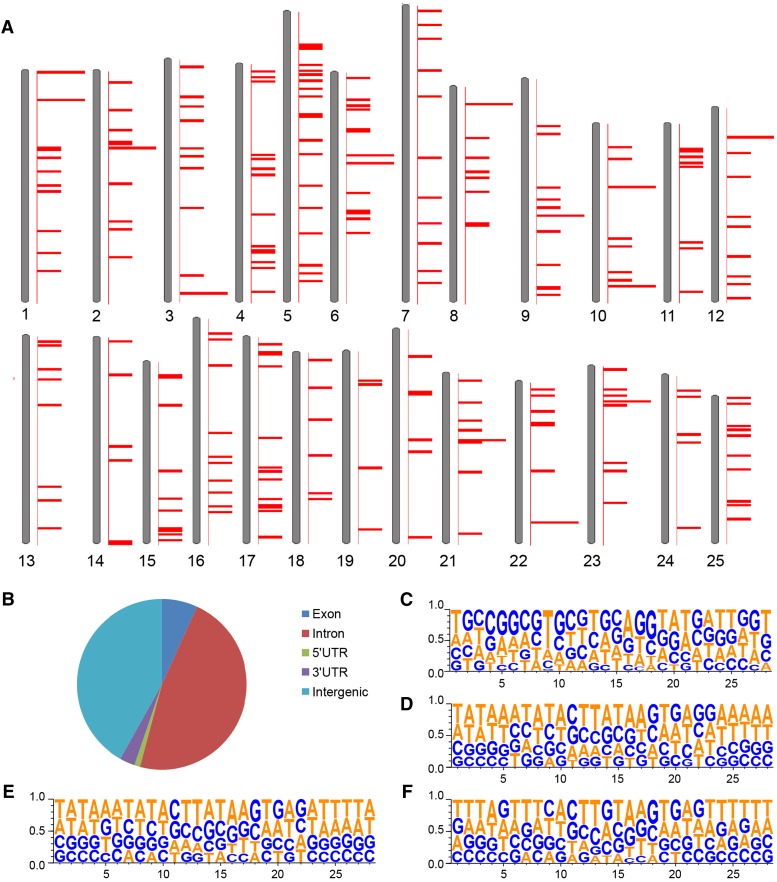
Figure 5.
Mapping of DsDELGT4 integrations to zebrafish chromosomes. Two hundred seventy-seven integrations sites (identified by TAIL-PCR from 283 founder lines) were used for this analysis. (A) Relative positions of integration sites on each chromosome according to zebrafish reference genome Zv9. (B) Distribution of insertions in specific regions of annotated genes (n = 277 insertions). (C–F) Alignment of 28 nucleotides around the integration sites located in intronic (C; n = 131), exonic (D; n = 30), or intergenic regions (E; n = 116) compared to all mapped Ds insertion sites (F). The eight nucleotides duplicated at integration sites are at nucleotide positions 11 to 18.