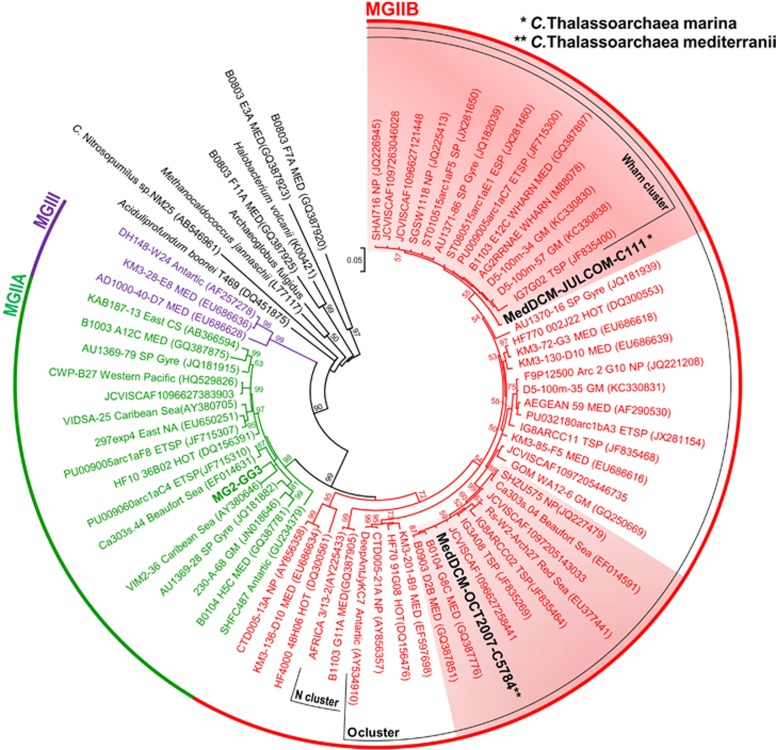
Figure 1.
16S rRNA phylogeny. Maximum-likelihood 16S rRNA gene tree (739 unambiguously aligned nucleotides) showing the relationship of the Thalassoarchaea (bold and black) with other MGII (MG2-GG3-assembled genome in bold and green). The clusters where thalassoarchaeal sequences appear are red shadowed. Numbers at nodes in major branches indicate bootstrap support (shown as percentages and only those >50%) by ML in MEGA 5.10. Scale bar represents the estimated number of substitutions per site. The different clusters are named following Galand et al., 2010. Sampling locations: MED, Mediterranean Sea; HOT, Hawaii Ocean Time-Series, North Pacific Gyre (ALOHA station); SP, South Pacific; ETSP, Eastern Tropical South Pacific; ESP, Eastern South Pacific; WP, western Pacific; SA, South Atlantic; NA, North Atlantic; GM, Gulf of Mexico; NP, North Pacific; CS, China Sea; NAEC, North American East Coast; GI, Galapagos Islands; CS, Caribbean Sea; TSP, Tropical South Pacific. The GOS data set identifiers are shown next to each GOS scaffold.