Abstract
Fifty-one Salmonella enterica serovar 4,[5],12:i:- (S. 4, [5],12:i:-) isolates (14 human strains, 34 animal strains and 3 river water strains) which are assumed to be monophasic variants of S. Typhimurium were analyzed using pulsed-field gel electrophoresis (PFGE) and multiple-locus variable-number tandem repeat analysis (MLVA) in order to investigate their genetic diversities and relationships. PFGE, MLVA and combination of them identified 28, 27 and 34 profiles (Simpson’s diversity indices [DI]=0.94, 0.96 and 0.97), respectively. No correlations were detected between MLVA clustering and PFGE clustering or phage typing. These results suggested that S. 4,[5],12:i:- originated from multiple S. Typhimurium ancestors. Two cattle and one pig isolates showing identical phage types as well as PFGE and MLVA profiles to human isolates S. 4,[5],12:i:- suggested the existence of the links between human infections and animal reservoirs.
Keywords: MLVA; monophasic variant; PFGE; Salmonella 4,[5],12:i:-(S. 4, [5],12:i:-); S. Typhimurium
The prevalence of Salmonella enterica serovar 4,[5],12:i:- among human salmonellosis has increased since the mid-1990s in many countries [5, 18]. In Japan, the sporadic cases caused by S. 4,[5],12:i:- occurred in Tohoku region since 2006 [21]. S. 4,[5],12:i:- is considered to be a monophasic variant of S. Typhimurium [4, 6, 7], and food-producing animals have been suspected as its reservoir [5, 8, 14]. However, the genetic relationship between human and animal isolates from Japan has not been investigated sufficiently.
To investigate genetic relatedness of microorganisms in detail, combined usage of multiple typing tools has been recommended [2]. Although PFGE has been considered as a gold standard for subtyping of Salmonella [16], several studies reported the less discrimination between highly genetically related strains [15]. Recently, Multiple-locus variable-number tandem repeat analysis (MLVA) has been proposed as an alternative tool to PFGE for molecular epidemiological analyses of S. Typhimurium [9, 13] and S. 4, [5],12:i:- [11, 12]. In the previous study, we analyzed 51 S. 4, [5],12:i:- isolates derived from humans and animal in Japan, using plasmid profile, antimicrobial susceptibility test and phage typing [10]. The result suggested that S. 4,[5],12:i:- isolates consisted of multiple distinct clones. In the present study, S. 4,[5],12:i:- isolates in Japan were characterized by PFGE and MLVA for the following purpose: (i) to verify usefulness of combined usage of PFGE and MLVA for typing of S. 4,[5],12:i:-, (ii) to clarify genetic diversity of isolates and (iii) to investigate the genetic relationship between human and animal isolates.
We examined 51 S. 4,[5],12:i:- isolates identified as monophasic variants of serovar Typhimurium in the previous study [10]. Fourteen isolates were obtained from sporadic cases of human salmonellosis. Seventeen were isolated from cattle with or without diarrhea, eight were isolated from asymptomatic swine or pork, 5 were isolated from asymptomatic poultry, 2 were isolated from asymptomatic crows, 2 were isolated from a penguin and parakeet diagnosed with salmonellosis, and 3 were isolated from river water. The epidemiological relationships were not found between the human cases and animal sources. The phage types of 3 isolates were as follows; 8 were DT193, 3 were DT26, 1 was DT27, 1 was DT120, 34 were RDNC, and 4 were untypeable (UT). PFGE was performed as described by Powell et al. [17]. Agarose-embedded chromosomal DNA was cleaved with BlnI (Roche Diagnostics, Basel, Switzerland) following the manufacturer’s instructions. Electrophoresis was performed in a 1% megabase agarose gel using the CHEF DR III apparatus (Bio-Rad Laboratories, Hercules, CA, U.S.A.) in 0.5× TBE (89 mM Tris, 89 mM boric acid and 2 mM EDTA) at 14°C, 6 V/cm. The pulse time was increased from 2.2 to 63.8 sec for 19 hr. A photograph of representative PFGE profiles was scanned and saved in TIFF format to be analyzed using Fingerprinting II software (Bio-Rad Laboratories). Digestion profiles of the isolates were compared each other by using Dice similarity coefficient. The cluster analysis was done using the hierarchic unweighted pair arithmetic average algorithm (optimization, 0.5%; tolerance, 1.0%), and a dendrogram was prepared. Profiles with minor variations (three or fewer band differences) were designed as different subtypes (indicated by different Arabic numerals) within a type (indicated by the same uppercase letters), which corresponded to similarities of approximately 80% or more [20]. Otherwise, profiles were designated as different genotypes (indicated by different uppercase letters) when rates of homology were less than 80%. MLVA was performed using 5 loci: STTR-9, STTR-5, STTR-6, STTR-10pl and STTR-3, as described previously [13]. DNA for PCR was isolated from cells scraped from Luria-Bertini (LB) agar plates, as described previously [19]. PCR amplification was performed using an initial denaturation cycle of 95°C for 5 min, followed by 30 cycles of 30 sec of denaturation at 94°C, 30 sec of annealing at 55°C and 1 min of extension at 72°C. Samples were then incubated for 3 min at 72°C to complete the extension. All PCR products were sequenced to confirm the number of tandem repeats. The amplified products were purified with QIAquick PCR purification columns (Qiagen, Hilden, Germany) and sequenced in both directions using Big Dye Terminator v3-1 chemistry (Applied Biosystems, Foster City, CA, U.S.A.). The number of tandem repeats at each locus was manually determined using Genetyx version 10.0 (Genetyx, Tokyo, Japan). The motif copy numbers present in the tandem array were imported into the BioNumerics software version 6.5 (Applied Maths, Sint-Martens-Latem, Belgium), and a minimum-spanning 3 (MST) was generated using the categorical coefficient. To evaluate the diversities of MLVA loci, Nei’s diversity indices were calculated using POPGENE version 1.32 (http://www.ualberta.ca/~fyeh/popgene.html).
To investigate the genetic relationship between human and animal isolates in detail, a combination usage of PFGE and MLVA was attempted. The isolates showing identical profiles both PFGE and MLVA were typed the same combination type indicated by Arabic numerals. To compare the discriminatory powers of PFGE, MLVA and combination of them, Simpson’s diversity index (DI) and 95% confidence intervals (CI) were calculated using Epicompare version 1.0 (http://www3.ridom.de/epicompare/).
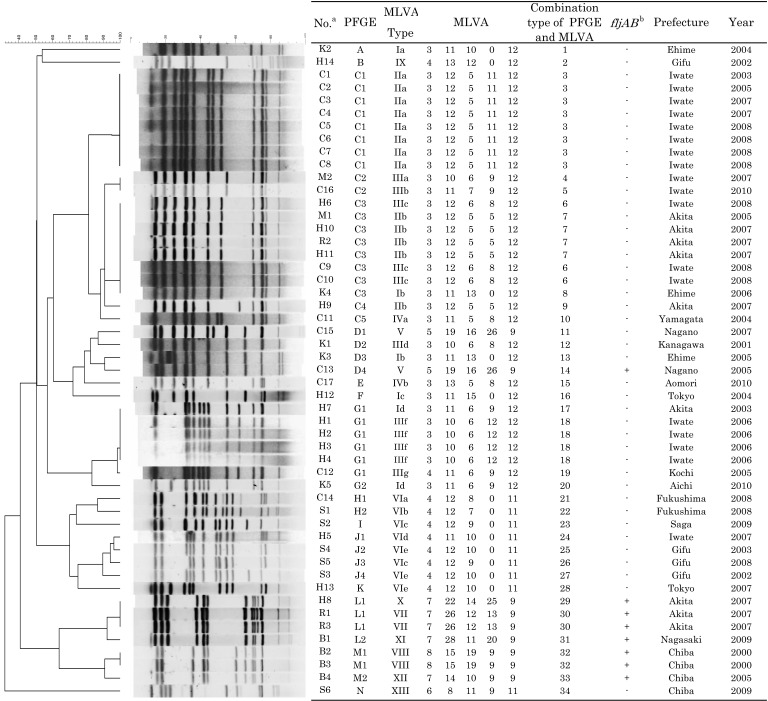
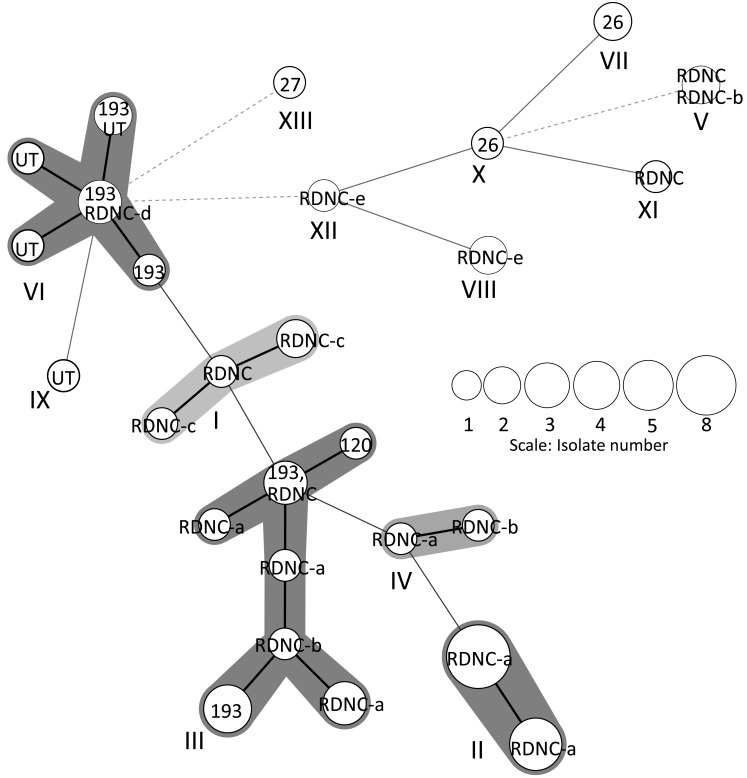
Figure 1 summarizes PFGE and MLVA typing of the 51 S. 4,[5],12:i:- isolates. Twenty-eight different PFGE profiles (DI, 0.94; 95% CI, 0.91–0.98) were identified by BlnI digestion. A cluster analysis of PFGE identified seven major clusters (C, D, G, H, J, L and M) and seven minor profiles (A, B, E, F, I, K and N). The dominant cluster C included 5 PFGE profiles (C1–C5) representing 20 isolates. The other clusters included between 2 and 4 PFGE profiles composed of between 2 and 7 isolates. MLVA identified 27 profiles (DI, 0.96; 95% CI, 0.93–0.98), and five MLVA loci varied in their degree of polymorphism, which was reflected by Nei’s diversity indices; STTR3, 0.52; STTR5, 0.74; STTR6, 0.83; STTR9, 0.58; STTR10pl, 0.85. A cluster analysis using MST revealed eight major clusters (I-VIII) and 5 minor profiles (IX-XIII), as shown in Fig. 2. The dominant cluster II included 2 MLVA profiles composed of 13 isolates. Cluster III included 7 MLVA profiles composed of 11 isolates. The other clusters included between 1 and 5 MLVA profiles composed of between 2 and 8 isolates. Three MLVA clusters, II, VII and VIII, included only one phage type. The other MLVA clusters included between 2 and 5 phage types, respectively.
Fig. 1.
Results of genotyping of S. 4,[5],12:i:- isolated from humans and animals in Japan. A dendrogram prepared from the PFGE profiles after BlnI digestion is placed on the left with their PFGE profiles. Detailed assignment of the results of PFGE and MLVA typing, epidemiological information and presence of fljAB operon [10] of the isolates are indicated in the table placed on the right. MLVA profiles are composed of five numbers indicating the repeat unit for each locus, in the following order: STTR9-STTR5-STTR6-STTR10pl-STTR3 [13]. aThe alphabets indicate the sources of isolates; C, cattle; H, human; S, swine; K, chickens; B, birds; M, pork; R, river water. bThe data were cited from the previous study [10].
Fig. 2.
Minimum-spanning tree of MLVA. Each MLVA type was indicated by nodes, displayed as circles, which were connected by branches of the minimum- spanning tree. The MLVA clusters differing by zero or one variable-number tandem loci were regarded as a group, highlighted and numbered by Roman numerals (I-XIII). A code within each circle indicated phage type. The length of the branches represented the genetic distances (changes in loci) between two neighboring types. The size of the circles depended on their population size. The thickness and dotting of the lines indicated the distance between the circles; a thicker line denoted a closer distance than a thin line, and a thin line denoted a closer distance than a dotted line.
Combining the PFGE and MLVA yielded 34 combination types (DI, 0.97; 95% CI, 0.94–0.99) (Fig. 1). The dominant combination type 3 (PFGE; C1, MLVA; IIa) was composed of the 8 bovine isolates obtained from different farms located in the same town. Combination type 6 (PFGE; C3, MLVA; IIIc) included 1 human and 2 cattle isolates derived from different towns. Combination type 7 (PFGE; C3, MLVA; IIb) included 2 isolates from different human sporadic cases, 1 isolate from a pig and 1 isolate from the sewage water of a different town. Combination type 18 (PFGE; G1, MLVA; IIIf) included 4 human isolates obtained from different sporadic cases in the same town. Combination type 30 (PFGE; L1, MLVA; VII) and 32 (PFGE; M1, MLVA; VIII) included 2 isolates each, which were obtained from different samples in the same town.
In the present study, various profiles were detected by PFGE and MLVA, respectively. The discriminatory power of combination typing was greater than single usage. These data suggest that combination typing of PFGE and MLVA is useful for discrimination of S. 4,[5],12:i:- isolates, as Kurosawa et al. reported previously [11]. On the other hand, the discriminatory power of phage typing (DI, 0.82; 95% CI, 0.74–0.91) of these isolates was lower than that of PFGE and MLVA [10]. Biased distribution of phage types seemed to reduce the DI value, and thus, we didn’t utilize phage typing data in this study.
Although the correlations between MLVA cluster, PFGE cluster and phage type were previously reported by the analyses of S. Typhimurium [13, 16], no correlation was found in this study. For example, MLVA cluster III included 5 distinct PFGE profiles (C2, C3, D2, G1 and G2). PFGE profile C3 was found in MLVA clusters II and IX. These results indicated that the isolates we examined consisted of multiple distinct clones. Previous studies suggested that S. 4,[5],12:i:- strains from several countries seem likely to be clonal. Most of S. 4,[5],12:i:- isolates from Germany were typed as DT193 [8]. In Spain and Thailand, U302 was the most prevalent phage type [1, 4]. However, Zamperini et al. [22] concluded that S. 4,[5],12:i:- isolates from Georgia were originated from multiple S. Typhimurium, since some monophasic isolates showed identical or similar PFGE profiles with S. Typhimurium. We identified that the deletion or point mutations in the fljAB operon and hin gene were the bases of the monophasic phenotype of the S. 4,[5],12:i:- isolates in the previous study [10]. In this study, 42 isolates lacking fljAB operon were divided into 15 clusters and 5 minor profiles by PFGE. MLVA divided these isolates into 6 clusters and 2 minor profiles. Eight isolates having the point mutations in the genes fljA and hin showed five PFGE profiles (D4, L1, L2, M1 and M2) and six MLVA profiles (V, VII, VIII, X, XI and XII). These results supported the hypothesis that S. 4,[5],12:i:- isolates in Japan were originated from multiple S. Typhimurium ancestors.
Best et al. [3] reported the presence of identical MLVA types across isolates from poultry, pigs and humans, suggesting that animals may be involved in the dissemination of S. Typhimurium through the food chain. Furthermore, several studies [8, 14] reported S. 4,[5],12:i:- isolates from pig, pork and humans showed identical or closely related profiles by PFGE or MLVA and suggested pig or pork might be a source of infection. In this study, two bovine isolates and one pig isolate belonging to combination types 6 and 7 showed identical phage types as well as PFGE and MLVA profiles to human isolates. Although, these strains were collected from the towns scattered throughout the prefecture, and there was no direct contact between these human and animals, these results suggest a possibility that food-producing animal is one of the reservoirs of S. 4,[5],12:i:- in Japan.
In conclusion, the present study demonstrated the usefulness of combined usage of PFGE and MLVA for S. 4,[5],12:i:- typing. The various profiles resulting from PFGE and MLVA suggested that S. 4,[5],12:i:- spreading in Japan may have originated from multiple S. Typhimurium ancestors. The presence of identical phage types as well as PFGE and MLVA profiles across human and animal isolates of S. 4,[5],12:i:- suggests the existence of the links between human infections and animal reservoirs.
Acknowledgments
We gratefully acknowledge the members of the prefectural livestock hygiene service centers and prefectural institutes of public health for kindly providing the Salmonella isolates used in this study. This work was partly supported by the Ministry of Health, Labour and Welfare of Japan (H24-Shokuhin-Ippan-008).
REFERENCES
- 1.Amavisit P., Boonyawiwat W., Bangtrakulnont A.2005. Characterization of Salmonella enterica serovar Typhimurium and monophasic Salmonella serovar 4,[5],12:i:- isolates in Thailand. J. Clin. Microbiol. 43: 2736–2740. doi: 10.1128/JCM.43.6.2736-2740.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbeit R. D.1995. Laboratory procedures for the epidemiologic analysis of microorganisms, pp. 190–208. In: Manual of Clinical Microbiology, 6th ed. (Murray, P. R., Baron, E. J., Pfaller, M. A., Tenover, F. C. and Yolken, R. H. eds.), American Society for Microbiology, Washington, D.C. [Google Scholar]
- 3.Best E. L., Lindstedt B. A., Cook A., Clifton Hadley F. A., Threlfall E. J., Liebana E.2007. Multiple-locus variable-number tandem repeat analysis of Salmonella enterica subsp. enterica serovar Typhimurium: comparison of isolates from pigs, poultry and cases of human gastroenteritis. J. Appl. Microbiol. 103: 565–572. doi: 10.1111/j.1365-2672.2007.03278.x [DOI] [PubMed] [Google Scholar]
- 4.de la Torre E., Zapata D., Tello M., Mejia W., Frias N., Garcia Pena F. J., Mateu E. M., Torre E.2003. Several Salmonella enterica subsp. enterica serotype 4,5,12:i:- phage types isolated from swine samples originate from serotype Typhimurium DT U302. J. Clin. Microbiol. 41: 2395–2400. doi: 10.1128/JCM.41.6.2395-2400.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Echeita M. A., Aladuena A., Cruchaga S., Usera M. A.1999. Emergence and spread of an atypical Salmonella enterica subsp. enterica serotype 4,5,12:i:- strain in Spain. J. Clin. Microbiol. 37: 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Echeita M. A., Herrera S., Usera M. A.2001. Atypical, fljB-negative Salmonella enterica subsp. enterica strain of serovar 4,5,12:i:- appears to be a monophasic variant of serovar Typhimurium. J. Clin. Microbiol. 39: 2981–2983. doi: 10.1128/JCM.39.8.2981-2983.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garaizar J., Porwollik S., Echeita A., Rementeria A., Herrera S., Wong R. M., Frye J., Usera M. A., McClelland M.2002. DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40: 2074–2078. doi: 10.1128/JCM.40.6.2074-2078.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hauser E., Tietze E., Helmuth R., Junker E., Blank K., Prager R., Rabsch W., Appel B., Fruth A., Malorny B.2010. Pork contaminated with Salmonella enterica serovar 4,[5],12:i:-, an emerging health risk for humans. Appl. Environ. Microbiol. 76: 4601–4610. doi: 10.1128/AEM.02991-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hopkins K. L., Maguire C., Best E., Liebana E., Threlfall E. J.2007. Stability of multiple-locus variable-number tandem repeats in Salmonella enterica serovar Typhimurium. J. Clin. Microbiol. 45: 3058–3061. doi: 10.1128/JCM.00715-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ido N., Lee K., Iwabuchi K., Izumiya H., Uchida I., Kusumoto M., Iwata T., Ohnishi M., Akiba M.2014. Characteristics of Salmonella enterica serovar 4,[5],12:i:- as a monophasic variant of serovar Typhimurium. PLoS ONE 9: e104380. doi: 10.1371/journal.pone.0104380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurosawa A., Imamura T., Tanaka K., Tamamura Y., Uchida I., Kobayashi A., Hata E., Kanno T., Akiba M., Yukawa S., Tamura Y.2012. Molecular typing of Salmonella enterica serotype Typhimurium and serotype 4,5,12:i:- isolates from cattle by multiple-locus variable-number tandem-repeats analysis. Vet. Microbiol. 160: 264–268. doi: 10.1016/j.vetmic.2012.05.023 [DOI] [PubMed] [Google Scholar]
- 12.Laorden L., Herrera-Leon S., Martinez I., Sanchez A., Kromidas L., Bikandi J., Rementeria A., Echeita A., Garaizar J.2010. Genetic evolution of the Spanish multidrug-resistant Salmonella enterica 4,5,12:i:- monophasic variant. J. Clin. Microbiol. 48: 4563–4566. doi: 10.1128/JCM.00337-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindstedt B. A., Vardund T., Aas L., Kapperud G.2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59: 163–172. doi: 10.1016/j.mimet.2004.06.014 [DOI] [PubMed] [Google Scholar]
- 14.Mossong J., Marques P., Ragimbeau C., Huberty-Krau P., Losch S., Meyer G., Moris G., Strottner C., Rabsch W., Schneider F.2007. Outbreaks of monophasic Salmonella enterica serovar 4,[5],12:i:- in Luxembourg, 2006. Euro Surveill. 12: E11–E12. [DOI] [PubMed] [Google Scholar]
- 15.Murphy T. M., McNamara E., Hill M., Rooney N., Barry J., Egan J., O’Connell A., O’Loughlin J., McFaddyen S.2001. Epidemiological studies of human and animal Salmonella typhimurium DT104 and DT104b isolates in Ireland. Epidemiol. Infect. 126: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ngoi S. T., Lindstedt B. A., Watanabe H., Thong K. L.2013. Molecular characterization of Salmonella enterica serovar Typhimurium isolated from human, food, and animal sources in Malaysia. Jpn. J. Infect. Dis. 66: 180–188. doi: 10.7883/yoken.66.180 [DOI] [PubMed] [Google Scholar]
- 17.Powell N. G., Threlfall E. J., Chart H., Rowe B.1994. Subdivision of Salmonella enteritidis PT 4 by pulsed-field gel electrophoresis: potential for epidemiological surveillance. FEMS Microbiol. Lett. 119: 193–198. doi: 10.1111/j.1574-6968.1994.tb06888.x [DOI] [PubMed] [Google Scholar]
- 18.Switt A. I., Soyer Y., Warnick L. D., Wiedmann M.2009. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i. Foodborne Pathog. Dis. 6: 407–415. doi: 10.1089/fpd.2008.0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamamura Y., Uchida I., Tanaka K., Okazaki H., Tezuka S., Hanyu H., Kataoka N., Makino S., Kishima M., Kubota T., Kanno T., Hatama S., Ishihara R., Hata E., Yamada H., Nakaoka Y., Akiba M.2011. Molecular epidemiology of Salmonella enterica serovar Typhimurium isolates from cattle in Hokkaido, Japan: evidence of clonal replacement and characterization of the disseminated clone. Appl. Environ. Microbiol. 77: 1739–1750. doi: 10.1128/AEM.01910-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., Swaminathan B.1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yatsuyanagi J., Waguri A., Sakuraba M., Aoki T., Kaneko N., Fujii S., Ohta M., Takahashi M., Kobayashi T., Ozawa N., Sugama K.2008. Serovars and antimicrobial susceptibilities of Salmonella isolates from sporadic diarrheal cases in Tohoku district, April 2006-March 2007. Infect. Agents Surveillance Rep. 29: 164–166(in Japanese). [Google Scholar]
- 22.Zamperini K., Soni V., Waltman D., Sanchez S., Theriault E. C., Bray J., Maurer J. J.2007. Molecular characterization reveals Salmonella enterica serovar 4,[5],12:i:- from poultry is a variant Typhimurium serotype. Avian Dis. 51: 958–964. doi: 10.1637/7944-021507-REGR.1 [DOI] [PubMed] [Google Scholar]