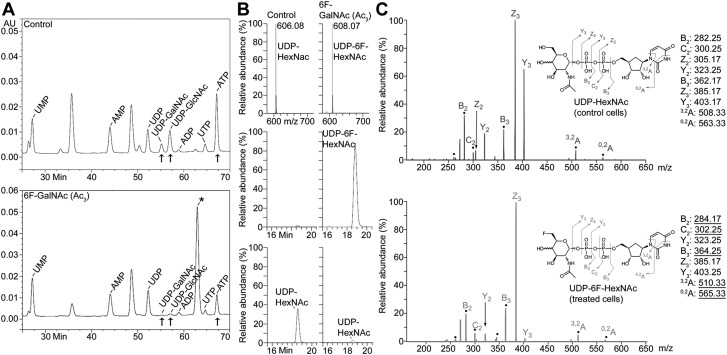
Figure 4.
6F-GalNAc (Ac3) is UDP activated and reduces cellular UDP-GlcNAc and -GalNAc. A) HPAEC-UV analysis of sugar nucleotides. An additional (sugar) nucleotide (*), most likely representing UDP-6F-HexNAc (see B), is present in 50 µM 6F-GalNAc (Ac3)-treated SKOV3 cells. Treatment of SKOV3 cells with 6F-GalNAc (Ac3) reduces UDP-GlcNAc, UDP-GalNAc, and ATP. The amount of 6F-GalNAc (Ac3)-treated cells used for extraction was 86% of the control. B) LC/MS analysis of sugar nucleotides. The mass of UDP-6F-HexNAc was increased by 2 amu, compared with UDP-HexNAc (top). The extracted ion current shows that UDP-6F-HexNAc was abundantly present after 50 µM 6F-GalNAc (Ac3) treatment (middle), whereas UDP-HexNAc (bottom) was strongly reduced. The amount of 6F-GalNAc (Ac3)-treated cells used for extraction was 79% of the control. C) Tandem MS with constellation of daughter ions of sugar nucleotides. UDP-HexNAc from control (untreated) cells (top) and UDP-6F-HexNAc from 6F-GalNAc (Ac3)-treated cells (bottom) were analyzed. Assignments and their m/z values are indicated by the molecular diagram (right). Ions marked with a dot represent NRE fragment ions that include carbon-6, which differs by 2 amu, depending on whether the parent molecule is HexNAc or 6F-HexNAc. A–C) The experiments shown were conducted together with those in untreated cells, for which the data have been published (20). MFI, mean fluorescence intensity; UMP, uridine 5′-monophosphate; UTP, uridine 5′-triphosphate.