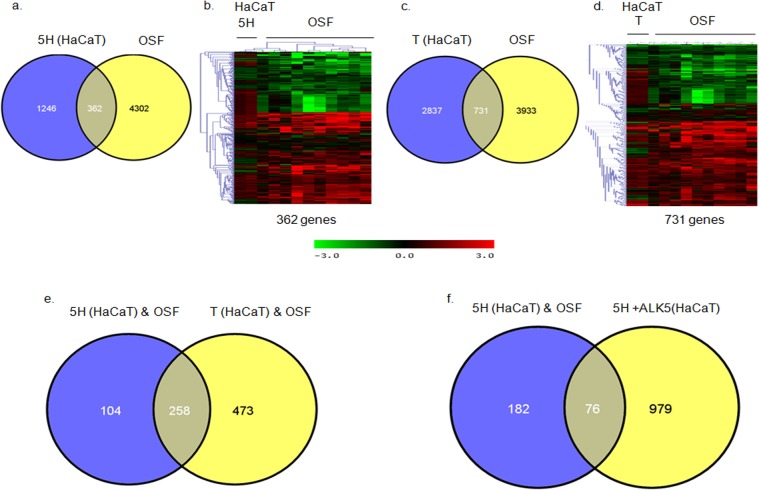
Fig 3. Genes regulated by areca nut in epithelial cells and OSF are via TGF-β.
Previously published transcriptome profile of OSF tissues (GSE 20170) [6] was compared with that of areca nut and/ or TGF-β regulated transcriptome in HaCaT (GSE 38227) [10] and hGF cells. a & b] Venn diagram and hierarchal cluster representing distribution of differentially or commonly regulated genes by areca nut in HaCaT cells and OSF. c & d] Venn diagram and hierarchal cluster representing distribution of differentially or commonly regulated genes by TGF-β in HaCaT cells and OSF. e] Venn diagram represents the 252 genes common between the 362 (252+104) genes regulated by areca nut in HaCaT and OSF and the 731 (252+ 473) genes regulated by TGF-β in HaCaT cells and OSF. f] Out of the 252 genes discussed in 3e, 182 genes are not regulated by areca nut in presence of ALK5 inhibitor (SB 431542) in HaCaT cells. This indicates that these 182 genes are regulated by areca nut are via TGF-β in HaCaT cells and possibly in OSF. In all hierarchal clusters; red, green and black colours represent up, down and un-regulated genes respectively. The rows represent genes and columns represent OSF and various treatments of areca nut (5H; 5 μg/ml) and/or TGF-β (T; 5 ng/ml).