Abstract.
The digital imaging and communications in medicine (DICOM) information model combines pixel data and its metadata in a single object. There are user scenarios that only need metadata manipulation, such as deidentification and study migration. Most picture archiving and communication system use a database to store and update the metadata rather than updating the raw DICOM files themselves. The multiseries DICOM (MSD) format separates metadata from pixel data and eliminates duplicate attributes. This work promotes storing DICOM studies in MSD format to reduce the metadata processing time. A set of experiments are performed that update the metadata of a set of DICOM studies for deidentification and migration. The studies are stored in both the traditional single frame DICOM (SFD) format and the MSD format. The results show that it is faster to update studies’ metadata in MSD format than in SFD format because the bulk data is separated in MSD and is not retrieved from the storage system. In addition, it is space efficient to store the deidentified studies in MSD format as it shares the same bulk data object with the original study. In summary, separation of metadata from pixel data using the MSD format provides fast metadata access and speeds up applications that process only the metadata.
Keywords: digital imaging and communications in medicine; multiseries digital imaging and communications in medicine; tag morphing; deidentification; attribute coercion; patient information reconciliation; order reconciliation; picture archiving and communication system, vendor neutral archive
1. Background and Significance
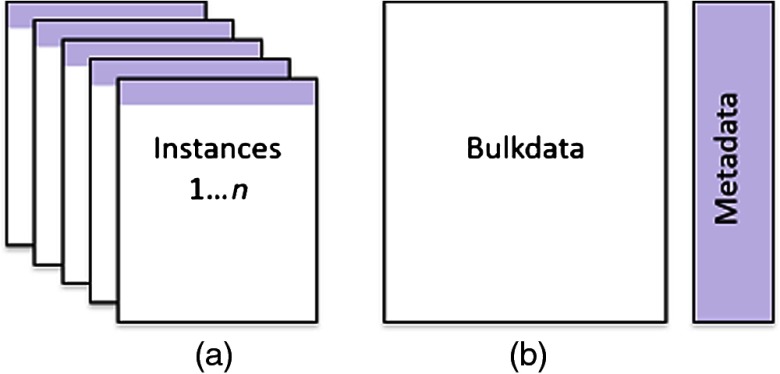
With the rapid development of three-dimensional imaging modalities such as computed tomography (CT) and magnetic resonance imaging (MRI), the size of DICOM studies is growing. These studies often contain hundreds of images. The standard DICOM format [see Fig. 1(a)] typically stores each image in a separate object, called an instance, which includes metadata and pixel data. The metadata contains information about the patient, the ordering physician, the imaging modality, frames of reference, scan parameters, image orientation, and so on. Some of these attributes are related to the study or series and do not vary with the image frames, e.g., the patient name, while others are image specific, e.g., the slice location. The size of the metadata is, in general, small compared to that of the pixel data. While the coupling of metadata and pixel data is reasonable for displaying individual images with related information, it creates unnecessary data redundancy, because the study and series level attributes are repeated with each object; in addition, many of the instance level attributes often have the same value across the series. Furthermore, combining pixel data and metadata in a single object increases the time required to update the metadata, because modifying the metadata also requires reading and possibly writing the pixel data, which is much larger than the metadata. Fast, easy access to metadata is desired for multiple real-world use cases, such as deidentification, order reconciliation, search indexing, and study migration across medical record number (MRN) domains. These applications rely on tag morphing, which is defined as retrieving a study from an archive and then adding, deleting or modifying one or more of the attributes of the study before transmitting or storing the modified study.1
Fig. 1.
Structure of: (a) a single frame DICOM (SFD) study versus that of (b) a multiseries DICOM (MSD) study.
Multiple modifications have been introduced to the DICOM standard that addressed the attribute redundancy issue and the coupling of the pixel data and metadata in one object. The enhanced multiframe object (MFD) was added to the standard to reduce metadata redundancy. It combines all instances in a series into one object. It uses an attribute called per-frame functional groups sequence that contains a sequence of datasets, where each dataset holds the attributes associated with a frame in the series. The advantage of MFD over traditional single frame DICOM (SFD) is that it does not repeat study and series level attributes within each instance in the series. However, the study level attributes are still replicated with each MFD object. Although the MFD format addressed the redundant series level attributes in DICOM studies, the coupling of the pixel data and its metadata still increases the processing time of the metadata.
Recently, the “composite instance retrieve without bulk data service” was added to the DICOM standard. This service allows instances to be retrieved without the attributes with large size values. The attributes defined in the service specification include the pixel data attribute, curve data attribute, overlay data attribute, and audio sample attribute.2 The service provides fast retrieval for metadata, however, applications that require metadata manipulation such as deidentification and study migration cannot benefit from this service. The reason is the service is added to facilitate the transmission of metadata between two application entities. The service is not concerned with updating the metadata in the storage system while both deidentification and study migration require the streaming of the whole study to a storage media after applying the metadata updates.
The one pass algorithm removes duplicated attributes from medical studies.3 The algorithm is efficient and has time complexity. Multiseries DICOM (MSD) is a format that enables fast access to DICOM metadata. MSD separates metadata from pixel data and uses the one pass algorithm to remove duplicated attributes.4 Figure 1 shows the difference between the traditional SFD and the MSD formats. For SFD, the DICOM study is represented by a set of instances, where each instance contains the relevant study, series and instance level attributes. Most, if not all, of the study and series attributes are duplicated in each instance. On the other hand, MSD combines all instances into two objects, metadata and bulk data. The metadata eliminates all unnecessary duplication of attributes. All attributes with value fields larger than 256 bytes are moved to the bulk data object, so that the metadata can be retrieved and parsed quickly.
In this work, the advantage of processing study metadata in MSD format from a file system is investigated. Two user scenarios that involve the manipulation of only the metadata are considered, study deidentification and study migration. The following subsections discuss them in detail.
1.1. Study Deidentification
The primary use of medical images is clinical; however, they are also used for research and teaching. DICOM objects are composed of different attributes including pixel data. Some of the attributes contain personally identifiable information.5 Those attributes are usually removed or modified prior to using the object for research or teaching. The DICOM standard defines the attributes that have to be removed and/or modified to deidentify instances.6 Table E.1-1 “application level confidentiality profile attributes,” DICOM part 15, lists all DICOM attributes tags and specifies for each attribute the action required for each security profile. Coupling metadata and pixel data has two limitations for deidentification. First, when the deidentified object is stored, the pixel data has to be replicated with the deidentified object even if there is no difference between the original and the deidentified pixel data. Second, the whole DICOM object including the pixel data has to be loaded into memory in order to deidentify it, which increases the processing time.
1.2. Study Migration
Some DICOM attributes have to be updated when a study is migrated between different entities to ensure that studies originating from one institution, or MRN domain, can be correctly imported into another MRN domain. Attributes such as patient name, patient ID, and issuer of patient ID must be updated to those of the new domain; other attributes such as issuer of accession number and accession number might be removed or modified, and the facility receiving the study might insert facility specific attributes.7 Migrating a large study stored in the traditional DICOM format requires many I/O operations (network and/or file system) to access potentially hundreds of SFD instances in order to update the metadata for every instance. Most picture archiving and communication system (PACS) and vendor neutral archives (VNAs), such as the DCM4CHEE archive, keep a copy of selected metadata attributes in a database.8 When these attributes need modification; only the database entry is updated, but not the files that contain the study. Then when the study files are retrieved, tag morphing is performed dynamically on each file in the study. This approach just delays the overhead of tag morphing until the study is retrieved, at which time the metadata in each file must be modified. It also makes the study metadata stored in the database inconsistent with that in the study files. This is undesirable because the database may become a bottleneck for study retrieval.
2. Materials and Methods
This work takes advantage of the MSD toolkit that was developed in previous work.3 The toolkit was built on top of the dcm4che2 toolkit,9 and contains a Java implementation of the one-pass deduplication algorithm. It supports reading and writing studies in SFD format and building a data model for the input study that is free of duplicated attributes. In this work, the toolkit is extended to support the MSD format. This section is organized as follows: Sec. 2.1 discusses the structure of the MSD format in detail. Section 2.2 contains detailed information about the dataset used. Sections 2.3 and 2.4 describe the design of the experiments for comparing the performance of the SFD and MSD formats for study migration and deidentification, respectively.
2.1. Multiseries DICOM Format
The MSD format was developed with three goals in mind: (1) aggregating all the instances of metadata in a study into one data structure, (2) separating the metadata from the bulk data, i.e., large values such as pixel, overlay and lookup table data, and (3) eliminating duplicate attributes. MSD is an extension of MFD, which combines all the images contained in a series into a single DICOM instance.
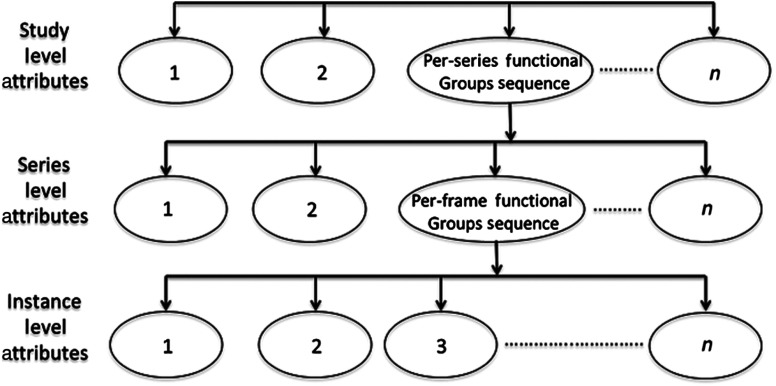
MSD extends this idea of attribute aggregation introduced by MFD from the series level to the study level. It uses a new attribute called per-series functional groups sequence to aggregate all the series level attributes into a single study object. This attribute contains a sequence of datasets, where each contains the attributes associated with a series in the study. These datasets have a similar structure to MFD. The study level attributes are stored once within the MSD object, thus are no longer duplicated with the series. Figure 2 shows the structure of the MSD metadata object. The one-pass deduplication algorithm is used to perform this aggregation by efficiently finding and removing repeated attributes. It actually reduces the overhead of parsing input studies.3
Fig. 2.
The nested structure of the MSD metadata.
The MSD format was developed to allow metadata and bulk data to be stored in separate objects. For this paper, bulk data is defined to be any attribute value larger than 256 bytes. The size of the bulk data object is limited to 1 GB. If the study size exceeds the maximum bulk object size, multiple bulk data objects are used. The study can have multiple bulk data objects even though the study size is less than 1 GB. This occurs if image frames are added to an existing study. A new bulk data object is created for each incremental study update. When a large value is moved into a bulk data object, the original attribute value is replaced by a bulk data reference. Data types in DICOM are known as value representations (VRs).10 For the MSD format, a new VR, with a symbol “BD,” was created for bulk data references. A bulk data reference contains 14 bytes that are structured as follows: The first two bytes contain the original VR of the attribute. The next four bytes hold the index of the bulk data object. This field identifies the bulk data object if the study has more than one bulk object. The following four bytes store the offset to the first byte of the attribute value within the bulk data object, and the last four bytes hold the size of the value. Figure 3 shows the representation of the pixel data attribute in the SFD format and its corresponding representation in the MSD format. MSD also addresses the limitation of current PACS and VNA implementations that store the metadata in a database and apply metadata changes to the database only (see Sec. 1.2).
Fig. 3.
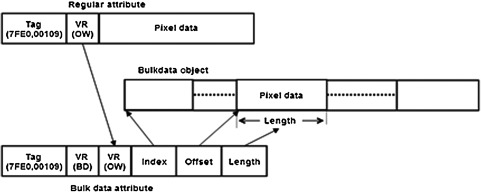
A pixel data attribute in traditional DICOM format and its counterpart in MSD format.
2.2. Input Dataset
The input dataset is composed of six different DICOM studies, three MRIs and three CTs. The study sizes range between 70 MB and 1.5 GB. All six studies are converted to MSD format. Both the original SFD studies and the converted MSD studies are used as input for the experiments described in the following sections. The dataset properties are shown in Table 1. The metadata is a small percentage of the overall study size. On average, the SFD metadata is 0.7% and the MSD is 0.16% of the original study size. The MSD toolkit was updated to include tag-morphing procedures, which are used for study migration and deidentification. The experiments were performed on a quad-core processor with 48 GB of physical memory and 8 GB of allocated heap memory.
Table 1.
Input study properties.
| Study name | Series | Images | Bulk data size (KB) | Metadata size | ||||
|---|---|---|---|---|---|---|---|---|
| SFD | MSD | |||||||
| (KB) | % | (KB) | % | SFD/MSD | ||||
| SMALLMR | 9 | 277 | 71,488 | 969 | 1.3% | 158 | 0.22% | 6.1 |
| SMALLCT | 5 | 338 | 173,088 | 920 | 0.5% | 174 | 0.10% | 5.3 |
| TESTMR | 17 | 1116 | 213,352 | 4278 | 2.0% | 594 | 0.27% | 7.2 |
| TESTCT | 7 | 1018 | 613,926 | 3310 | 0.5% | 1193 | 0.19% | 2.8 |
| TESTCTA | 13 | 2524 | 1,366,321 | 8052 | 0.6% | 2845 | 0.21% | 2.8 |
| BREASTMR | 22 | 2362 | 1,499,589 | 8605 | 0.6% | 1285 | 0.09% | 6.7 |
| Average | 12 | 1273 | 656,294 | 4356 | 0.7% | 1041 | 0.16% | 4.2 |
2.3. Study Migration Experiment
This experiment was designed to assess the time required to process SFD and MSD metadata for study migration. The processing time required to apply tag morphing to the input dataset for manipulating the attributes associated with study migration is recorded. The following steps are carried out on each of the twelve studies: (1) read each study from the file system into memory, (2) update the values of a predefined set of attributes in the study with dummy values, (3) save the updated study from memory to the file system in its original format (SFD or MSD), and (4) record the time for steps 1 to 3. The chosen attributes to be updated are issuer of patient ID, patient ID, and accession number. It is mandatory to update those attributes when a study is transferred across MRN domains.
2.4. Study Deidentification Experiment
Application program interfaces (APIs) for deidentifying study’s metadata were added to the MSD toolkit. They implement the basic application level confidentiality profile defined in part 15 appendix E of the DICOM standard.6 For each study in the dataset, both the time required to deidentify the study and the space required to store the deidentified version were recorded. Verifications tests were developed to ensure that the studies are deidentified properly according to the standard. They also verify that the patient identity was removed and deidentification method code sequence attributes are updated to mark the study as deidentified and list the deidentification options used, respectively.
3. Results
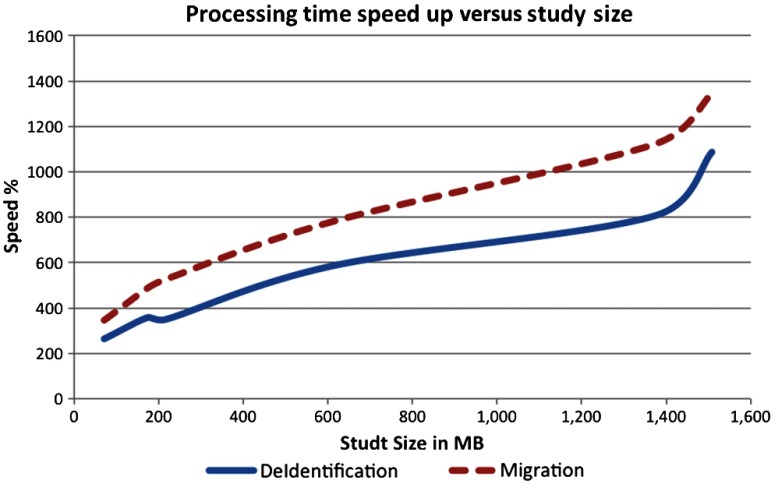
The results in Table 2 show that the processing time for migrating the studies stored in MSD format is, on average, more than eight times faster than applying the same changes to those in the standard SFD format. A similar observation can be made from the results of the deidentification experiment in Table 3. The time required to deidentify the studies stored in MSD format is almost six times less than the time required to deidentify them in SFD format. Figure 4 shows the relationship between the study size and the speedup achieved. Sizes of the deidentified studies are recorded in Table 4. It is evident that, in terms of storage, the overhead of deidentifying MSD is negligible compared to deidentifying SFD.
Table 2.
Study migration performance, SFD versus MSD.
| Study name | SFD time (ms) | MSD time (ms) | Speedup (%) |
|---|---|---|---|
| SMALLMR | 519 | 150 | 346 |
| SMALLCT | 791 | 162 | 488 |
| TESTMR | 1518 | 288 | 527 |
| TESTCT | 2282 | 291 | 784 |
| TESTCTA | 4695 | 418 | 1123 |
| BREASTMR | 5251 | 390 | 1346 |
| Average | 2509 | 283 | 886 |
Table 3.
Study deidentification performance, SFD versus MSD.
| Study Name | SFD Time (ms) | MSD Time (ms) | Speedup (%) |
|---|---|---|---|
| SMALLMR | 631 | 239 | 264 |
| SMALLCT | 935 | 261 | 358 |
| TESTMR | 1482 | 423 | 350 |
| TESTCT | 2597 | 442 | 588 |
| TESTCTA | 5165 | 640 | 807 |
| BREASTMR | 6044 | 556 | 1087 |
| Average | 2809 | 427 | 576 |
Fig. 4.
Migration/deidentification speedup versus study size.
Table 4.
Study deidentification overhead, SFD versus MSD.
| SFD total (KB) | De-Id SFD (KB) | % Overhead | MSDtTotal (KB) | De-Id MSD (KB) | % Overhead |
|---|---|---|---|---|---|
| 72,457 | 72,112 | 99.5% | 71,646 | 78 | 0.11% |
| 174,008 | 173,646 | 99.8% | 173,262 | 80 | 0.05% |
| 217,630 | 215,858 | 99.2% | 213,946 | 310 | 0.14% |
| 617,236 | 615,946 | 99.8% | 615,119 | 372 | 0.06% |
| 1,374,373 | 1,371,475 | 99.8% | 1,369,166 | 901 | 0.07% |
| 1,508,194 | 1,504,964 | 99.8% | 1,500,874 | 692 | 0.05% |
| 660,650 | 659,000 | 99.6% | 657,336 | 406 | 0.08% |
4. Discussion
There are two reasons for the significant improvement achieved by processing DICOM metadata in MSD format rather than the traditional SFD format. First, the MSD metadata is deduplicated, which makes its size four times smaller, on average, than the metadata size in SFD format (see Table 1), and consequently reduces its parsing time. Second, and more importantly, the MSD format does not need to load the bulk data into memory or write it back to the file system, which reduces I/O time and memory footprint. In addition, it should be noted that performance of MSD improves as the study size increases. As shown in Fig. 4, the speedup is proportional to the study size. This performance difference is especially important for large hospitals that process hundreds of imaging studies daily. It could be even more important for regional health information exchanges that will eventually manage billions of images for millions of people.
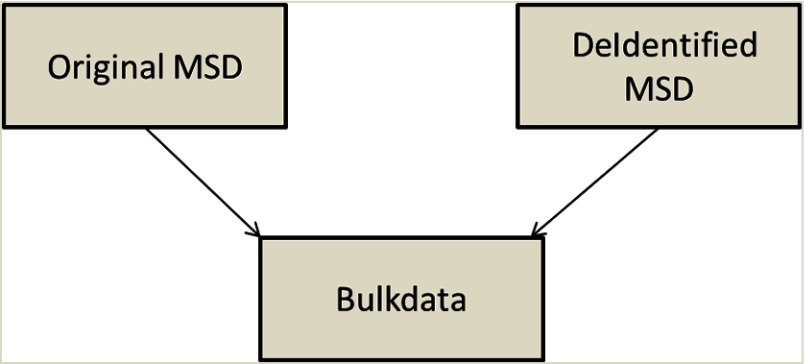
The results show the size of the deidentified studies equals to its metadata size. There is no need to store and duplicate the bulk data object if there is no patient information burned with the pixel data. Both the original and the deidentified studies can reference and share the same bulk data object; see Fig. 5.
Fig. 5.
Deidentified MSD studies storage. The original and deidentified objects share the same bulk data object.
5. Conclusion
The MSD format is a novel format for medical images storage that separates metadata from bulk data and thus allows operations that only access metadata to be significantly faster. The results show a significant improvement in metadata processing time when study is stored in MSD format rather than SFD format. MSD is efficient for applications that include tag morphing such as deidentification and study migration for two reasons. The primary reason is that there is no need to read or transmit the bulk data, such as pixel data, to access the metadata. Second, the size of MSD metadata is significantly smaller than that of the traditional SFD metadata. Moreover, MSD based storage systems address the limitation of current PACS and VNA implementations that store the metadata in a database and apply metadata changes only to the database. With MSD, the PACS database and images stored in the archive are kept in sync with respect to each other. To our knowledge, this is the first published work with actual measurements to demonstrate the value of separating DICOM format metadata from bulk data.
Future work includes an evaluation of the end-to-end scenario that models the common tag morphing used in cases in clinical facilities. The framework used in this experiment reads and writes the studies from the file system while, in reality, the studies are stored in a PACS’s archive and transferred to a remote workstation on demand. Accessing studies from the archive introduces transmission delays. The end-to-end scenario where a study is retrieved from a PACS’s archive, updated, and transmitted over the network to a remote workstation will show the performance of MSD-based PACS versus SFD ones in a real clinical workflow.
Acknowledgments
The authors would like to thank “Mr. Yu Ning” for his effort and contribution toward developing the MSD toolkit. The toolkit has API for retrieving DICOM studies and exporting them as MSD files. The toolkit is the foundation for achieving this work. We are also grateful for Mr. Ning’s comments on the manuscript that improved the final version of this work.
Biographies
Mahmoud Ismail is a PhD student in the Computer Science Department, Johns Hopkins University. His area of research is medical imaging informatics. He has got two masters degrees in 2009 from the Systems and Biomedical Engineering Department, Cairo University, and in 2011 from the Computer Science Department, Johns Hopkins University. In addition to his research experience, he has a wide industrial experience gained through working for multiple corporations, including Mentor Graphics, Google, Microsoft, and currently Amazon.
James Philbin is a co-director of the Center for Biomedical and Imaging Informatics, Johns Hopkins University. He is an accomplished executive with more than 20 years of experience as a scientist and entrepreneur. He is an acknowledged expert in image management, data architecture, high-performance computing, and infrastructure design. He is the co-chair of DICOM Work Group 27 and has been or is on medical advisory boards for Amicas, Emageon, Siemens, and Vital Images.
References
- 1.DeJarnette W. T., “Context management and tag morphing in the real world,” White Paper Series, 2010, http://www.dejarnette.com/downloads/get.aspx?i=45048 (March 2015).
- 2.DICOM Standard, Digital Imaging and Communications in Medicine (DICOM), Part 4: Service Class Specifications, NEMA, Rosslyn, VA (2015).
- 3.Ismail M., Philbin J., “Multi-series DICOM: an extension of DICOM that stores a whole study in a single object,” J. Digital Imaging 26 691–697 (2013). 10.1007/s10278-013-9577-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ismail M., Philbin J., “Fast, storage efficient de-identification of medical studies,” in Proc. The DICOM Int.Conf. and Seminar, Bangalore: (2013). [Google Scholar]
- 5.McCallister E., Grance T., Scarfone K., Guide to Protecting the Confidentiality of Personally Identifiable information, National Institute of Standards and Technology, Special Publication 800-122, Gaithersburg (2010).
- 6.DICOM Standard, Digital Imaging and Communications in Medicine (DICOM), Part 15: Security and System Management Profiles, NEMA, Rosslyn, VA (2013).
- 7.Van Ooijen P. M. A., et al. , “Incorporating out-patient data from CD-R into the local PACS using DICOM worklist features,” J. Digital Imaging 18(3), 196–202 (2005). 10.1007/s10278-005-5158-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.dcm4chee home page, “dcm4che,” [online], http://www.dcm4che.org/confluence/display/ee2/Home (12 October 2014).
- 9.dcm4che, “dcm4che toolkit,” [online], http://dcm4che.org/confluence/display/d2/dcm4che2+DICOM+Toolkit. (February 2014).
- 10.National Electrical Manufacturers Association, Digital Imaging and Communications in Medicine (DICOM) Part 6: Data Dictionary, National Electrical Manufacturers Association, Rosslyn, VA (2011).