Fig 2. Pedigrees and PHEX gene mutation chromatograms from two Indian families with X-linked hypophosphatemic rickets.
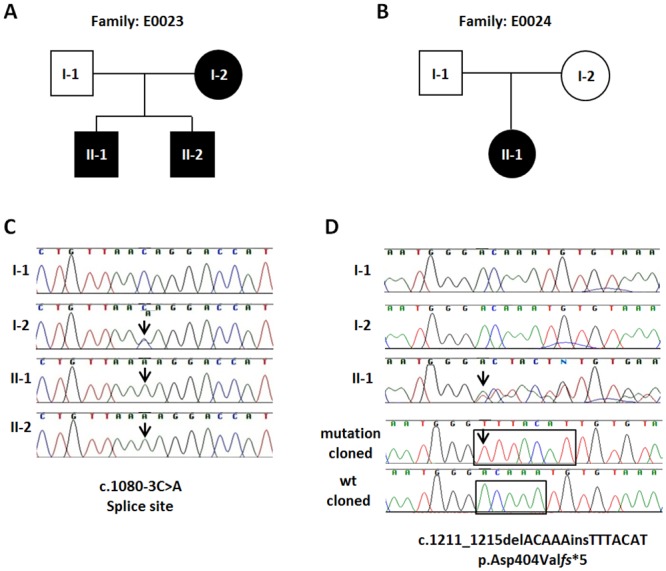
(A,B) Pedigrees of two XLH families. Filled symbols represent affected individuals. Circles and squares indicate females and males, respectively. (C) Genomic sequence chromatograms showing a novel acceptor splice site mutation c.1080-3C>A in intron 9 of the PHEX gene (arrows). The mutation in family E0023 co-segregates with the affected status and is present in the mother (I-2) and her two affected sons (II-1 and II-2). The father (I-1), who is healthy, exhibits a hemizygous wild-type allele. The mutation was identified after whole exome sequence analysis in E0023-II-2 and confirmed by direct Sanger sequencing. (D) A novel heterozygous insertion/deletion mutation (c.1211_1215delACAAAinsTTTACAT) in PHEX exon 11 leading to a frameshift (p.Asp404Valfs*5) was identified by direct Sanger sequencing in a female patient with hypohosphatemic rickets (E0024-II1). Note, that both her father (I-1) and her mother (I-2) show only the wild-type sequence. Therefore, we conclude that this mutation is most likely a de novo change, although the presence of germ line mosaicism in one of the parents cannot entirely be ruled out by analyzing RNA from blood only. The two bottom chromatograms represent the c.1211_1215delACAAAinsTTTACAT frameshift mutation (bottom) and the wild-type allele (above) after the respective patients’ PHEX exon 11 PCR products have been cloned into TA-cloning plasmid vector pCR2.1 (Invitrogen) and subsequently sequenced. Black boxes highlight deleted (ACAAA) and inserted (TTTACAT) bases accordingly.
