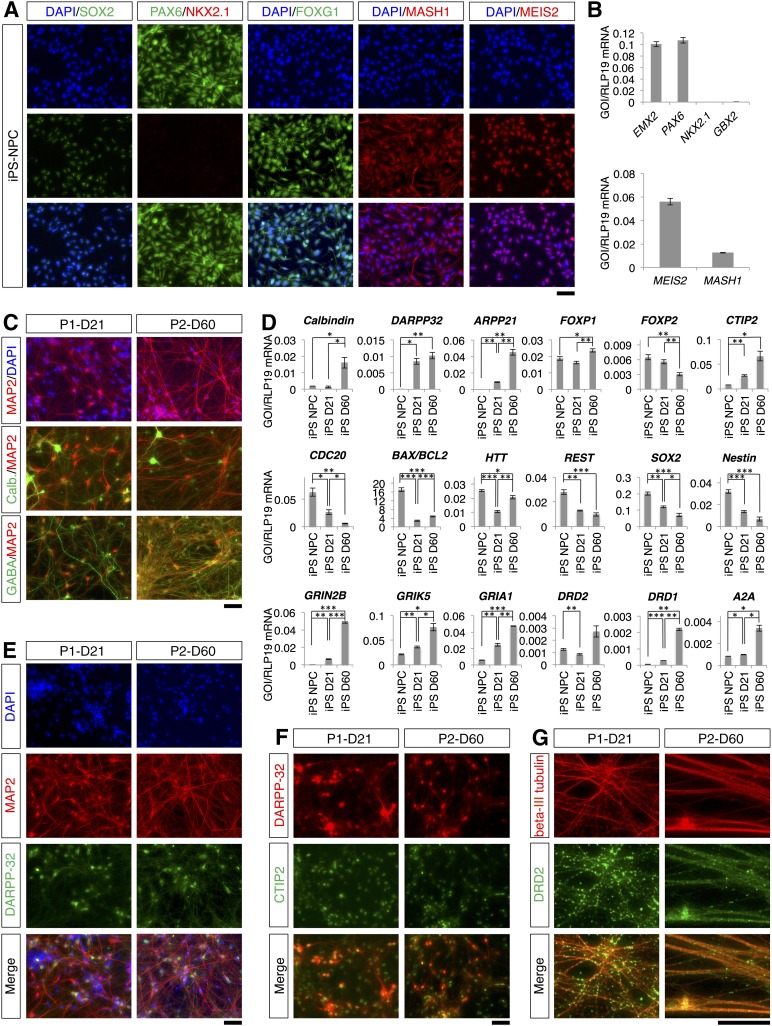
Figure 7.
Differentiation of striatal projection neurons from iPS-derived NPCs. (A): iPS-derived NPCs immunostained for SOX2, PAX6, NKX2.1, FOXG1, MASH1, and MEIS2. (B): Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis of EMX2, PAX6, NKX2.1, GBX2, MEIS2, and MASH1 expression. (C): Colocalization of MAP2 with calbindin and with GABA at the endpoint of differentiation. (D): qRT-PCR analysis of marker expression in iPS-derived NPCs and differentiated cells. Levels of statistical significance were set at ∗, p < .05, ∗∗, p < .01, and ∗∗∗, p < .001. (E): Colocalization of MAP2 and DARPP-32 at the end of the differentiation protocols. (F): Colocalization of DARPP-32 and CTIP2 at the end of the differentiation protocols. (G): Colocalization of β-III tubulin and DRD2. The scale bars correspond to 50 μm on the specimen level. Abbreviations: D, day; DAPI, 4′,6-diamidino-2-phenylindole; IPS, induced pluripotent stem cell; NPC, neural progenitor cell; P1, two-step protocol; P2, three-step protocol.