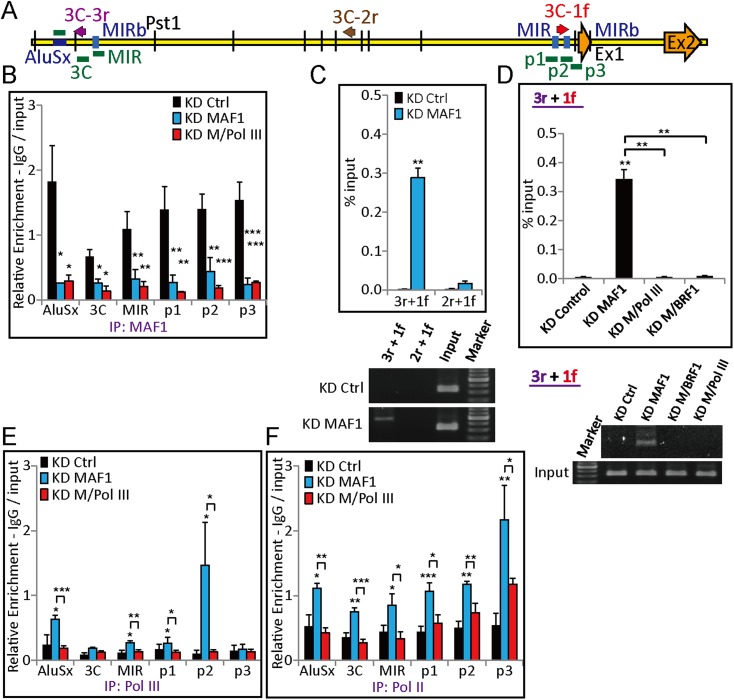
Figure 7. Pol III is required for chromatin looping at the GDF15 promoter after MAF1 knockdown.
(A) Schematic diagram of GDF15 with ChIP–qPCR amplicons (AluSx, 3C, MIR, p1, p2, and p3), the orientation of 3C primers (arrows: 3C-3r, 3C-2r, and 3C-1f), and locations of exons (Ex1 and Ex2). (B) ChIP with anti-MAF1 antibody (IP: MAF1) was performed in MCF-7 cells subjected to siRNA knockdown of MAF1 (KD MAF1) or simultaneous knockdown of MAF1 and Pol III (KD M/Pol III) for 72 hr. Binding of MAF1 was detected at the GDF15 promoter, which diminished after MAF1 knockdown. (C) A 3C assay was performed as indicated in the ‘Materials and methods’, and DNA was subjected to PCR. Chromatin looping was detected after MAF1 knockdown from 3C-3r to 3C-1f (top panel) and is shown by a representative gel (bottom panel). (D) The induced chromatin looping after MAF1 knockdown (KD MAF1) was diminished when MAF1 underwent simultaneous knockdown with either Pol III (KD M/Pol III) or BRF1 (KD M/BRF1) (top panel) and is shown by a representative gel (bottom panel). (E) ChIP with anti-Pol III antibody (IP: Pol III) or anti-Pol II antibody (IP: Pol II) was performed in MCF-7 cells subjected to siRNA knockdown. Enhanced binding of Pol III was detected at the GDF15 promoter after MAF1 knockdown, which was abolished when there was simultaneous knockdown of Pol III and MAF1 (KD M/Pol III). (F) MAF1 knockdown indicates enhanced binding of serine 5-phosphorylated Pol II, which was abolished when there was simultaneous knockdown of Pol III and MAF1. All data shown represent the mean ± s.e.m., n ≥ 3, *p < 0.05, **p < 0.01, ***p < 0.001 (t-test).