Figure 2.
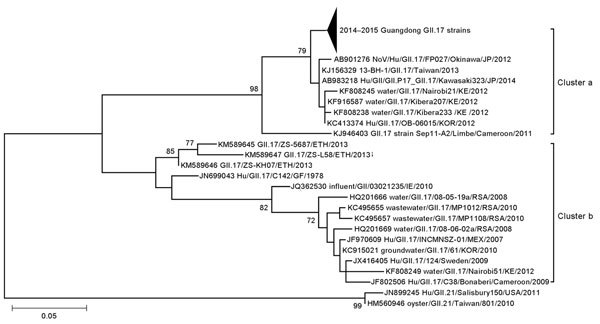
Phylogenetic tree of noroviruses based on the 282-bp region of the capsid N terminus/shell gene. Nucleotide sequences were analyzed by using the maximum-likelihood method. Supporting bootstrap values >70 are shown. The subtrees of GII.17 detected in Guangdong Province, China, during 2014–2015 were compressed. GII.21 genotype strains were used as outgroups. Scale bar indicates nucleotide substitutions per site. Sequences of 24 reference norovirus strains are included. Arrowhead represents number of strains from Guangdong, 2014–2015. ETH, Ethiopia; GF, French Guiana; IE, Ireland; JP, Japan; KE, Kenya; KOR, Korea; RSA, The Republic of South Africa; MEX, Mexico.
