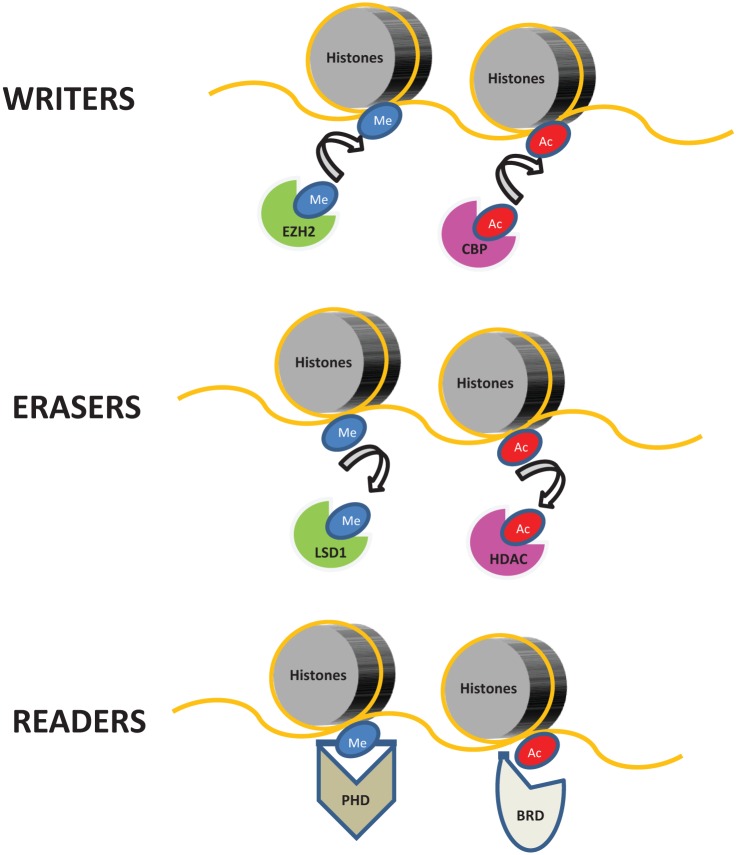
Figure 1.
Classification of epigenetic regulators.
Epigenetic regulators can be broadly divided into three groups. Epigenetic writers, such as methyltransferases [e.g. enhancer of zest homolog 2 (EZH2)] and acetyltransferases [e.g. CREB binding protein (CBP)] are enzymes responsible for covalently modifying histones by adding methyl (Me) or acetyl (Ac) groups to specific lysine residues. Epigenetic erasers, such as demethylases [e.g. lysine specific demethylase 1 (LSD1)] and histone deacetylases (HDAC), catalytically remove these histone marks. Finally, epigenetic readers are proteins which can recognize and selectively bind to specific covalent modifications of histones using highly specialized protein domains, such as plant homeodomain (PHD) finger and bromodomain (BRD). It should be noted that DNA does incur similar functional modifications, although this is not represented in the figure.