Fig. 1.
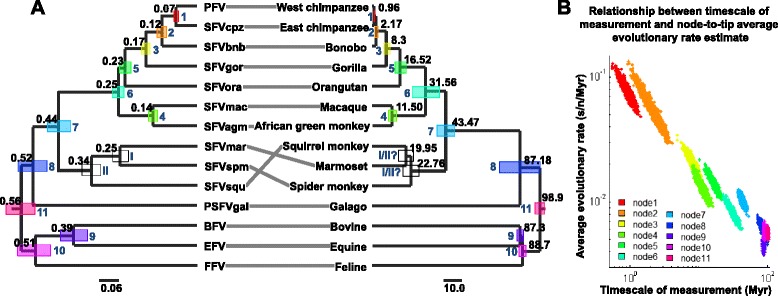
Foamy virus (FV) and corresponding host phylogenies, and the relationship between node-to-tip average evolutionary rate estimate and timescale of measurement. (a, left) FV phylogeny (taxon definitions and GenBank sequence accession numbers are in Additional file 1: Table S1). Black numbers are estimated total per-lineage nucleotide substitutions in the units of substitutions per site. The node bars represent the uncertainties of the estimated nucleotide substitution divergences. The scale bar is in units of substitutions per site. (a, right) Host tree. Black numbers are estimated divergence dates in units of millions of years, for which the estimation uncertainties are shown by node bars. The scale bar is in units of millions of years. The topology of the host tree and the divergence dates were estimated elsewhere (see the references in Additional file 1: Table S2). Solid lines between the two trees indicate FV-host associations, and blue Arabic numbers (1-11) indicate matching FV-host nodes (i.e. FV-host co-speciation events). Nodes within the FV tree that could not be mapped conclusively onto the host tree are labelled by blue Roman numbers (I and II). The colours of the node bars on the FV tree correspond to the colours of the bars on the host tree. b Timescales of rate measurement and node-to-tip average evolutionary rate estimates on a log-log scale. The node numbers (1-11) refer to those in the FV and host trees (a). The colours correspond to the node bars’ colours. The summary of the raw data can be found in Additional file 1: Table S2
