Figure 1. Pathway analysis of up- and down-regulated genes in hepatocellular carcinoma.
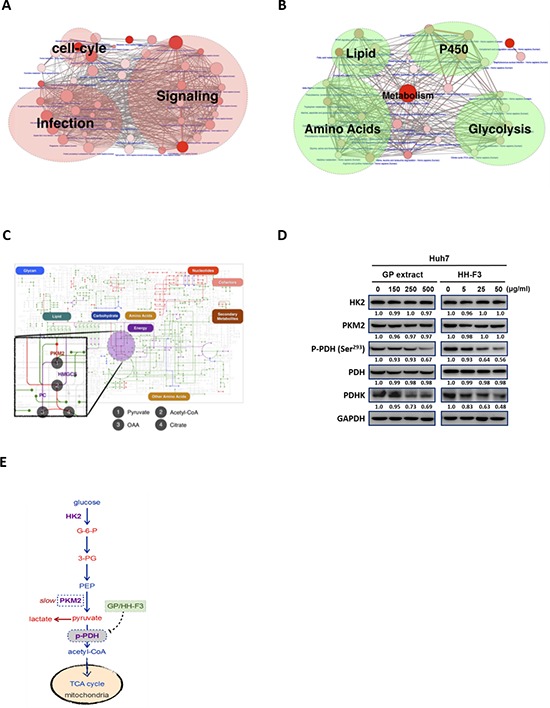
Two network diagrams are presented here to illustrate the pathways enriched by up- and down-regulated genes, respectively. (A) Up-regulated genes were mainly enriched in signaling, infection, and cell cycle-related pathways, whereas (B) down-regulated genes were enriched only in metabolism pathways related to lipid synthesis, glycolysis, amino acid metabolism, and P450 metabolism. Each node represents a pathway, and the number of genes in the pathway determines the size of the node. The larger the pathway, the bigger the size of the node is. The P-value from pathway enrichment analysis determines the redness of the balls. The lower the p-value, the redder the node is. The thickness of the lines between nodes in the network represents the number of genes that the two pathways have in common. (C) The blueprint of metabolism pathways adapted from KEGG to illustrate dysregulated reactions based on EHCO3 (green lines: reactions whose enzymes are down-regulated; red lines: reactions whose enzymes are up-regulated). Purple lines are reactions whose enzymes in HCC could be reversed from down-regulation to up-regulation by GP/HH-F3, whereas blue lines are reactions whose enzyme expression levels could be suppressed by GP/HH-F3. (D) Huh7 cells were treated with 30% DMSO GP extracts and HH-F3 for 24 h, respectively. Western blot shows the effects after GP/HH-F3 treatment for HK2, PKM2, pyruvate dehydrogenase complex (PDH), phosphorylated PDH (p-PDH) and PDHK with GAPDH used as a control. (E) GP/HH-F3 could influence the TCA cycle via down-regulation of phosphorylated PDH, but GP/HH-F3 could not affect other molecules in the TCA cycle. A possible mechanism describing how GP/HH-F3 restores dysregulated glycolysis. Glucose-6-phosphate (G-6-P), 3-phosphoglycerate (3-PG), and phosphoenolpyruvate (PEP) are key intermediates in glycolysis.
