Figure 2. Schematic overview of the bile acid transport inhibition module in DILIsym.
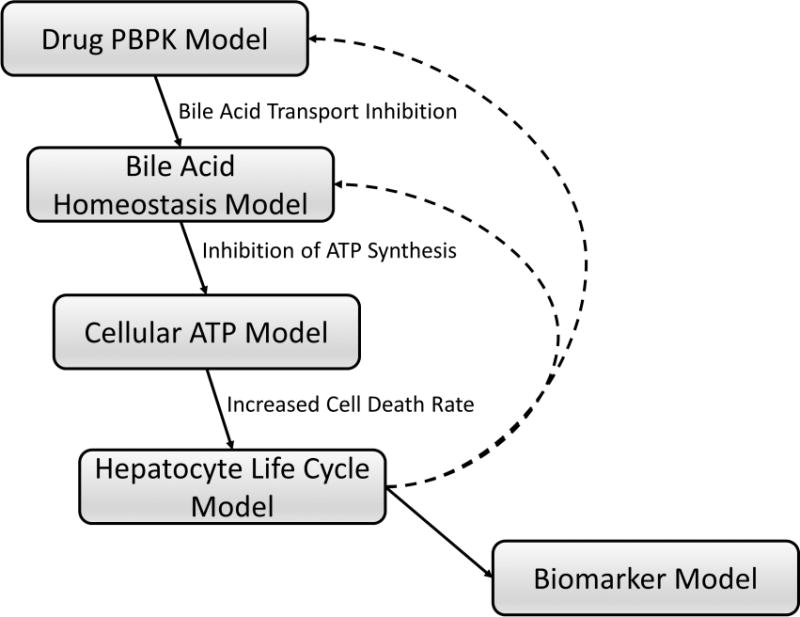
Hepatic and systemic disposition of drugs/metabolites are simulated using a physiologically-based pharmacokinetic (PBPK) model (Drug PBPK Model). The Bile Acid Homeostasis Model represents hepatobiliary disposition and enterohepatic recirculation of lithocholic acid (LCA) and chenodeoxycholic acid (CDCA) species, and all other (bulk) bile acids.20 Using bile acid transport inhibition constants of drugs/metabolites (e.g., Ki, IC50), altered bile acid disposition is simulated. Increased hepatocellular accumulation of bile acids inhibits hepatic ATP synthesis and decreases intracellular ATP concentrations (Cellular ATP Model), leading to necrotic cell death (Hepatocyte Life Cycle Model) and elevations in serum biomarkers of hepatocellular injury and function (e.g., ALT, AST, bilirubin) (Biomarker Model). Loss of hepatocytes will subsequently influence drug and bile acid disposition (dashed lines), allowing dynamic interaction between kinetics and toxicity mechanisms. Details regarding the construction and structures of sub-models can be found in the supplementary materials.
