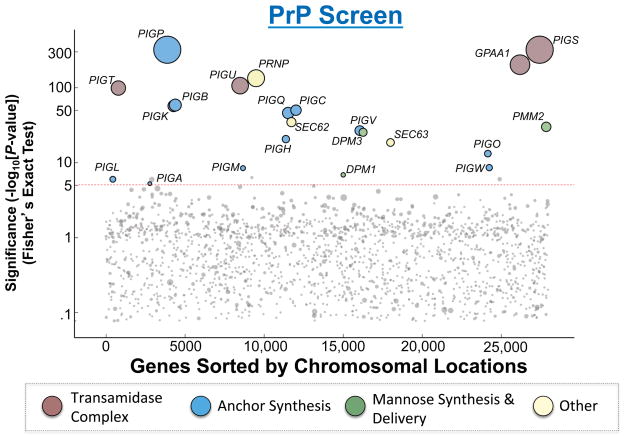
Figure 2. Hits from the haploid genetic screen of the PrP pathway.
The Y-axis represents the −Log10 of P values for the gene hits in the selected population as compared to a published unselected control (Jae et al., 2013) using Fisher’s exact test. We set a P value cutoff of 1×10−5 to account for multiple hypothesis testing. In addition, for genes with a P value less significant than 1×10−10, we only considered genes as hits if they also had strong bias for sense-strand intron insertions. The X-axis represents the chromosomal positions of the genes. Circle size is scaled according to the number of unique inactivating gene-trap insertions a gene received. Circles are colored according to functional groups. Dashed line indicates the cutoff of significance.