Figure 5. GSNL-1 acts downstream of CED-3 and can be cleaved by CED-3.
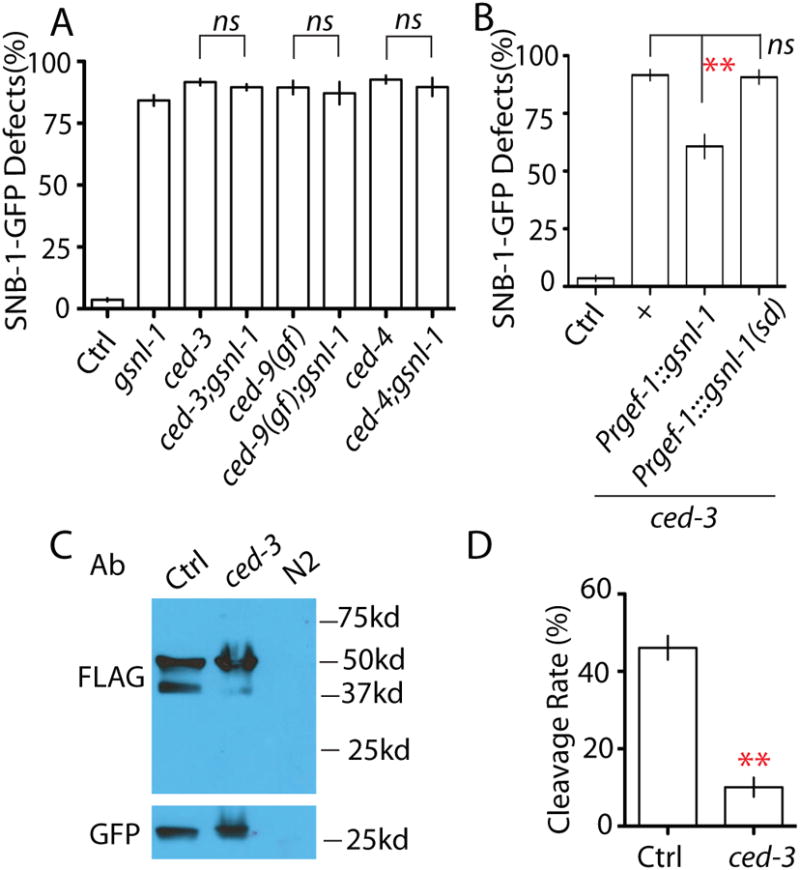
(A) gsnl-1 acts in the same pathway with CED genes. Double mutants of gsnl-1 and CED genes have similar degree of SNB-1-GFP elimination defects as single mutants. (B) The F-actin severing function of gsnl-1 is required for synapse elimination induced by activation of the cell death pathway. Overexpression of gsnl-1 can partially suppress ced-3 phenotypes in RME neurons, and deleting the linker region required for severing function abolishes its suppression ability on ced-3(lf). (C-D) CED-3 can cleave GSNL-1. (C) Western blotting analysis shows loss of function in ced-3 suppresses the cleavage of GSNL-1. FLAG antibodies recognize the full length (50kd) and the cleaved (40kd) GSNL-1. No signal was detected in wild type N2 worms without the transgene. Pttx-3∷gfp was used as a co-injection marker for gsnl-1 transgene, and GFP was served as a loading control in western blot. (D) Quantification of cleavage rate of GSNL-1. Five independent experiments were performed, and band intensity was measured using ImageJ. The cleavage rate was calculated by: P40 Intensity/(P50 intensity+ P40 intensity) × 100%. In Figure A and B, experiments were performed at least 3 times, with N ≥80 animals each time. For transgenic animals the results shown here are generated from at least three independent lines. Data is shown as mean ± SD. Student test, ** P < .01, ns: no significant difference.
