Figure 6. CED-3 cleaves GSNL-1 at a C-terminal conserved site to promote its F-actin severing ability.
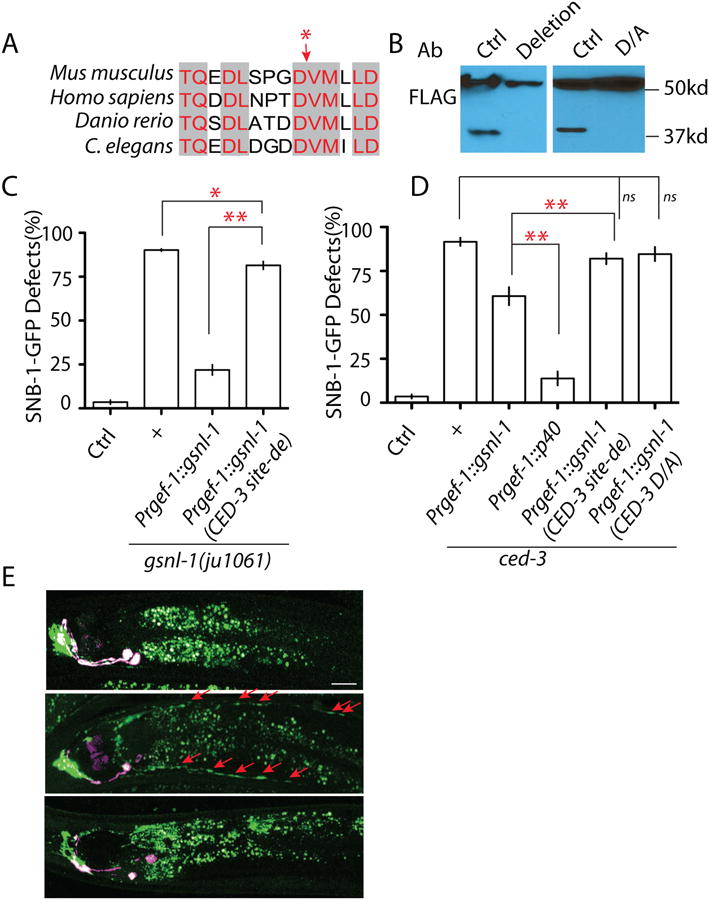
(A) Sequence analysis identifies a conserved candidate caspase cleavage site at the region where cleavage can produce a 40kb band in C. elegans. NCBI Reference Sequence: Danio rerio XP_003198825.1; Mus musculus (gene: Advillin) XP_006513165.1; Homo sapiens (Gene: Advillin) NP_006567.3. The red star points the potential cut site. (B) Deletion of the five core amino acids “DDVMI” (deletion) or mutating the aspartate (“DDVMI”) to alanine (“DAVMI”) (D/A) in this region blocks the cleavage of GSNL-1. (C) Deletion of the CED cleavage site strongly affects gsnl-1 functions. Expression of gsnl-1 lacking the CED cleavage site only shows weak rescue ability, while wild type gsnl-1 rescues mutant phenotypes in over 90% animals (D) Expression of the cleaved GSNL-1 can bypass the requirement of ced-3 in synapse elimination. Expression of P40 almost totally suppresses ced-3 phenotypes, while un-cleavable GSNL-1 fails to suppress ced-3 phenotypes. (E) Loss-of-function in the CED pathway induces mis-accumulation of F-actin in D/V neurites, and overexpression of P40 can suppress the effect of ced-4(lf). In Figure C and D, experiments were performed at least 3 times, with N ≥80 animals each time. For transgenic animals the results shown here are generated from at least three independent lines. Data is shown as mean ± SD. Student test, ** P < .01, * P<.05, ns: no significant difference.
