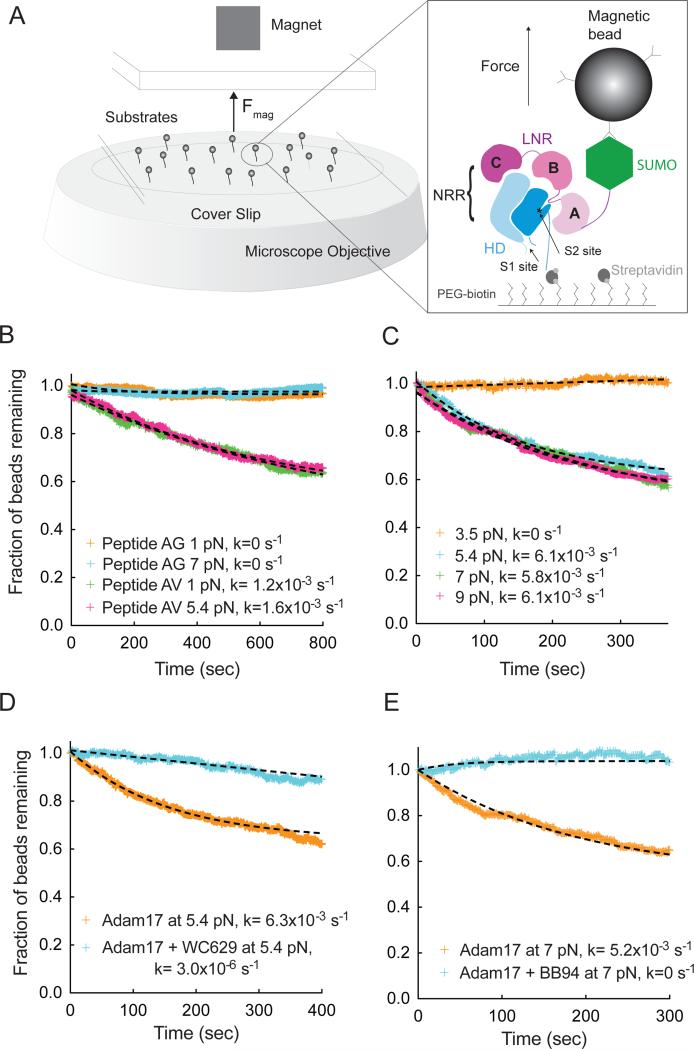
Figure 1. Single molecule assay of Adam17-mediated proteolysis of the Notch1 NRR under force.
(A) Assay schematic and experimental design. Magnetic beads are tethered to proteins immobilized in a flowcell mounted on an inverted microscope. Force is applied to the beads by varying the distance between a magnet and the surface of the flowcell. Substrate proteolysis is monitored by determining the fraction of beads released over time. The expanded view in the right-hand panel illustrates the Notch1 NRR, captured on the flow cell with streptavidin and tethered to the magnetic bead using anti-SUMO antibodies. (B) Adam17-catalyzed proteolysis of biotinylated and SUMO tagged recombinant peptides, containing either the natural S2-cleavage site sequence (AV, green and pink), or a mutated sequence with a V1721G substitution (AG, orange and cyan) at the forces indicated. (C) Adam17-catalyzed proteolysis of the Notch1 NRR, monitored as a function of time at different levels of applied force. Traces shown represent averages of 2 or 3 replicates. (D, E) Effect of inhibitors on proteolysis of the Notch1 NRR in the single molecule cleavage assay. Traces shown represent a single experiment. (D) Effect of WC629, an anti-Notch1 inhibitory antibody that binds to the NRR, on the time course of Adam17-catalyzed NRR proteolysis. (E) Effect of BB94, an ADAM inhibitor, on the time course of Adam17-catalyzed NRR proteolysis. Additional control experiments are provided in Figure S1.