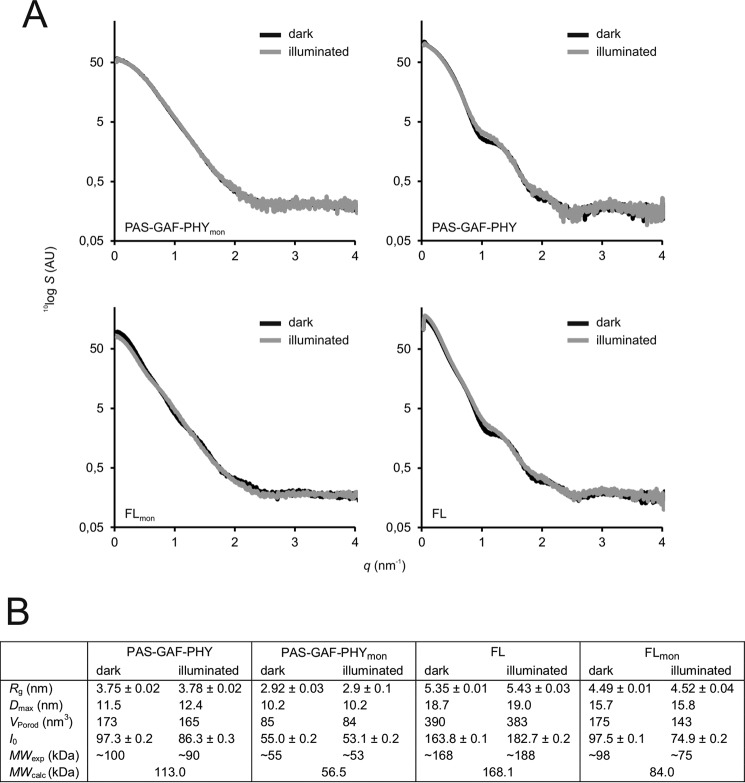
FIGURE 4.
SAXS of the phytochrome constructs. A, scattering of the phytochrome fragments (S) as a function of scattering angle (q) is plotted on a logarithmic scale. Dark (Pr) state samples are plotted as black lines; illuminated (mostly Pfr) state samples are plotted in gray lines. The scattering data of PAS-GAF-PHY is from Takala et al. (11). B, structural parameters calculated from the SAXS scattering. Rg, radius of gyration; Dmax, maximum particle dimension; VPorod, Porod volume; I0, forward scattering. The molecular weights (MWexp) were estimated from the I0 of BSA using the formula MWexp ≈ MW(BSA) × [I0(sample)/I0(BSA)], where MW(BSA) = 66 kDa. Theoretical molecular weights (MWcalc) were calculated from the protein sequence (58).