Extended Data Figure 3.
Related to Figure 2
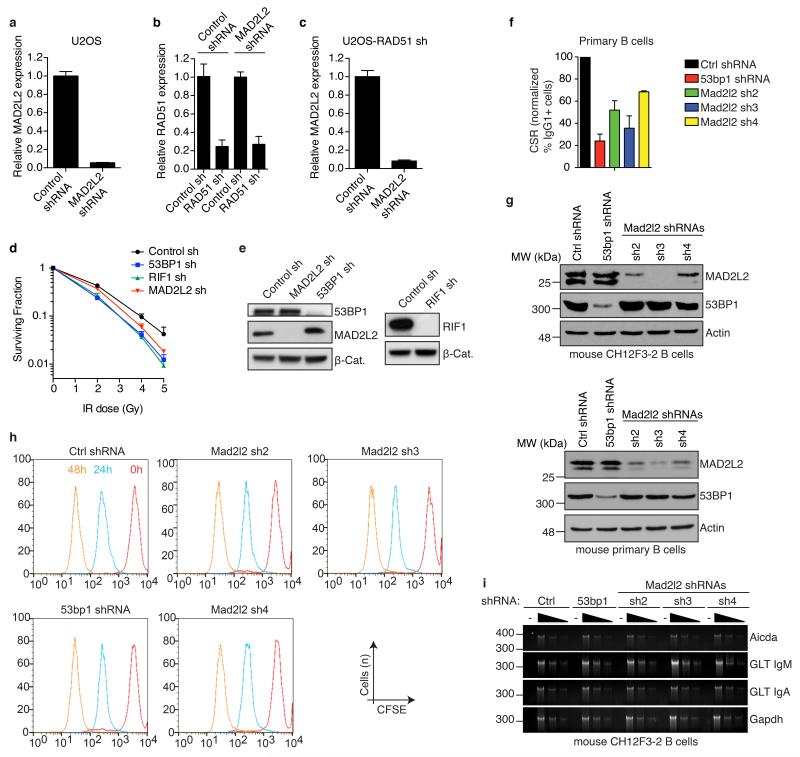
a, qRT-PCR analysis of MAD2L2 expression levels in U2OS cells infected with control or MAD2L2 shRNAs, and used in the repair assays shown in Fig. 2d (Error bars: s.d.). b, c, qRT-PCR analysis of RAD51 (b) and MAD2L2 (c) expression levels in RAD51-depleted, E6E7-expressing U2OS cells used in the assays shown in Fig. 2e (Error bars: s.d.). d, Clonogenic survival assays of U2OS cells transduced with non-targeting control or 53BP1, RIF1 or MAD2L2 shRNAs and treated with the indicated doses of IR (n=3-4, ± s.e.m.). e, Western blot analysis of 53BP1, MAD2L2 and RIF1 in U2OS cells transduced with the indicated shRNAs. f, CSR in shRNA-transduced primary B cells (n=2, ± s.d.). g, Western blot analysis of MAD2L2 and 53BP1 in CH12F3-2 B cells and mouse primary B cells transduced with the indicated shRNAs. h, MAD2L2 depletion does not affect cellular proliferation in murine B cells. CH12F3-2 cells transduced with control, 53bp1 or Mad2l2 shRNAs were loaded with CFSE and analysed at 0, 24 and 48 h post-stimulation by flow-cytometry. Profiles from all time points are plotted in the same histogram. i, MAD2L2 depletion does not affect the transcription of critical genes implicated in CSR. RT-PCR analysis of AID (Aicda) mRNA, IgM (GLT IgM) and IgA (GLT IgA) germline transcript levels using 2-fold serial dilutions of cDNA made from activated CH12F3-2 B cells transduced with the indicated shRNAs. Gapdh was used as a control for transcript expression.