Fig. 1.
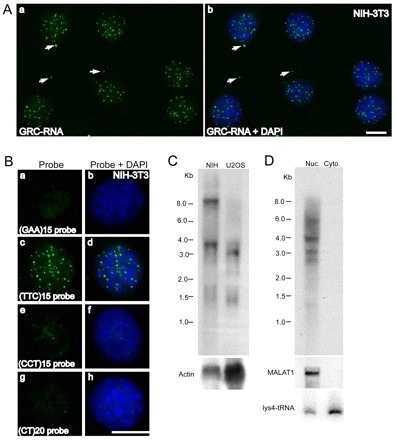
GRC-RNA is a member of the nrRNA family of lncRNAs. (A) RNA-FISH using an AK082015 probe (green) in NIH-3T3 cells revealed the punctate nuclear localization of GRC-RNA. In addition to nuclear foci, GRC-RNA also localized to a few cytoplasmic foci (arrows). Scale bar: 10 μm. (B) RNA-FISH using FITC-labeled (GAA)15 (a,b), (TTC)15 (c,d), (CCT)15 (e,f) and (CT)20 (g,h) single-stranded oligonucleotide probes in NIH-3T3 cells revealed that only the (TTC)15 probe hybridized to nuclear-enriched GRC-RNA (c,d). DNA was counterstained with DAPI (blue). Scale bar: 10 μm. (C) Northern blot using a 32P-labeled (TTC)15 probe from total RNA revealed that GRC-RNAs represent heterogeneous transcripts. Actin RNA was used as a loading control. (D) Northern blot using a 32P-labeled (TTC)15 probe in nuclear and cytoplasmic RNA fractions from EpH4 cells revealed nuclear enrichment of GRC-RNA. MALAT1 and lysine tRNA were used as markers for nuclear and cytoplasmic RNA fractionation, respectively.
