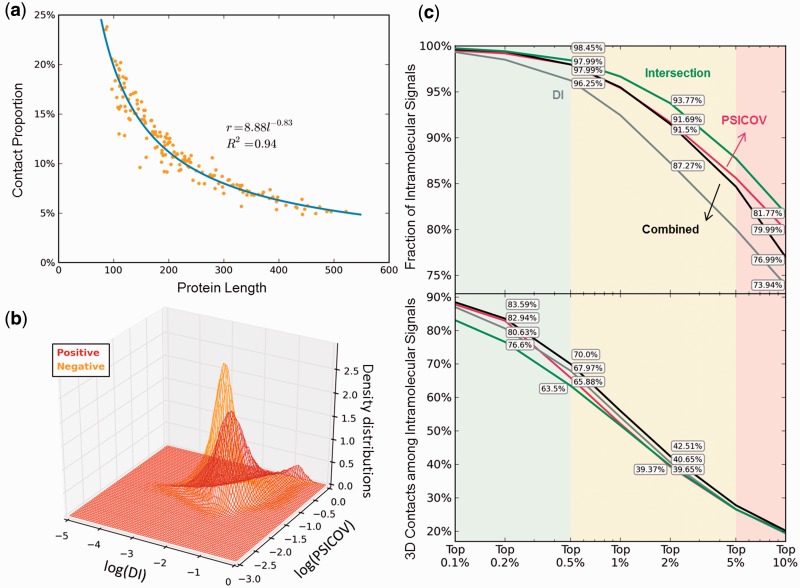
Fig. 7.
Development of hybrid methods. (a) Assessment of prior probability of 3D contact, P(+), by a regression analysis of a training set of 162 structurally known protein sequences. (b) Density distributions of positive and negative signals, P(DI, PSICOV∣+) and P(DI, PSICOV∣−) (see Equation 1), modelled by kernel density estimation. (c and d) Comparative performance of the individual methods DI (gray) and PSICOV (red), and the combined naïve Bayes classifier method (Equation 1) (black), based on the fraction of intramolecular signals (c) and fraction of 3D contact-making pairs (d). The predictions based on the intersection of MIp, DI and PSICOV are shown by the green curve