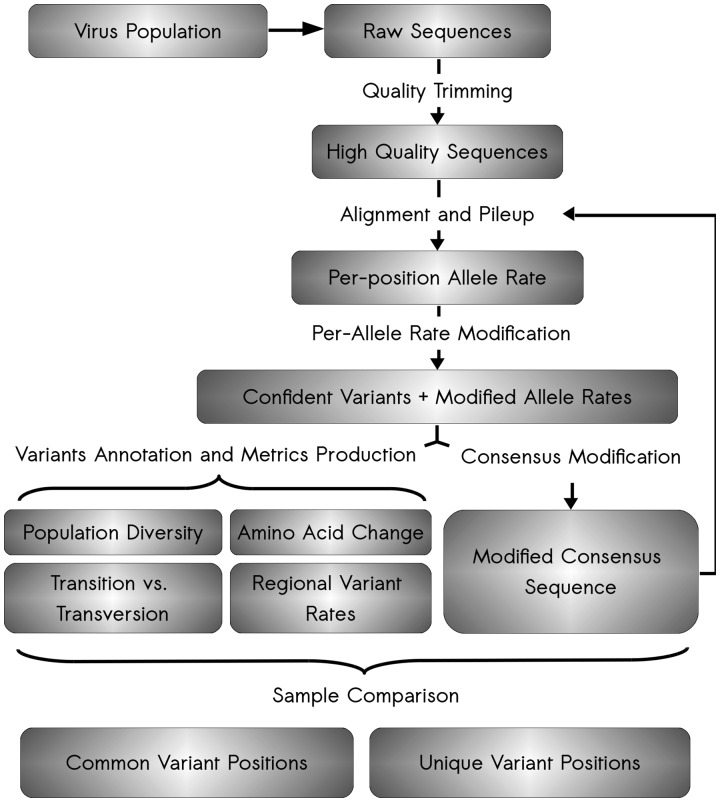
Fig. 1.
Schematic of ViVan pipeline workflow. The analysis starts with raw sequence reads output by deep sequencing of a virus population sample. First, these raw reads undergo quality trimming where low quality bases are removed from both ends of the read. Second, these quality reads are aligned against a user-supplied reference sequence and a pileup is produced for each position. The pileup output is then analyzed, true variants are identified, variant frequencies are modified and confidence intervals calculated. From these modified significant variants, an assortment of variation metrics is produced, including information regarding the predicted amino acid change in each protein, the variation rates across the viral genome, transition/transversion rates and specific nucleotide change tables. Additionally, once variant frequencies have been calculated, a consensus sequence is produced, utilizing the major allele in each position. This modified consensus sequence can then be used for the alignment of the initial quality reads, hence improving overall alignment and accuracy. Once the analysis is done for each virus sequence sample, a comparison is performed between groups of samples in order to pinpoint both common and unique variants in each group