Fig. 4.
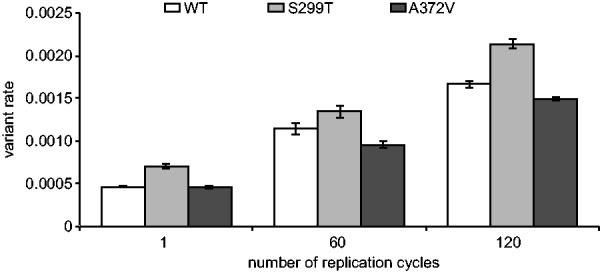
Temporal accumulation of variants found in viral populations. The total number of minority variants found in each CVB3 strain (wild type WT, high fidelity A372V and low fidelity S299T) was calculated for each sequenced passage (1, 60 and 120 replication cycles). Variation rates (the average proportion of variant alleles across covered bases in the genome sequence) ± SEM are shown, from three biological replicates, (P < 0.0001)
