Fig. 4.
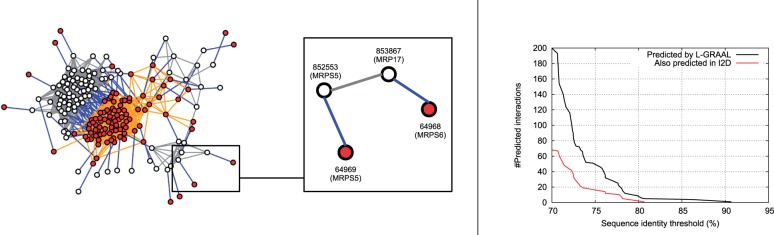
Predicting new protein interactions. Left: Part of L-GRAAL's alignment that aligns human and yeast ribosome pathways. The PPI sub-network of yeast (white nodes and gray edges) is mapped to the PPI sub-network of human (red nodes and orange edges) as indicated by the blue edges. The inset highlights a predicted interaction: Proteins MRPS5 and MRP17, which are interacting in the yeast PPI network, are aligned to proteins MRPS5 and MRPS6, which are not interacting in the human PPI network. Right: Using the whole L-GRAAL's alignment between yeast and human PPI networks, we plot in black the number of predicted interactions (y axis) as a function of the minimum sequence identity between the aligned yeast-human proteins (x axis). We add in red the number of these predicted interactions that are also predicted in I2D database
