Table 1. List of parameters used to describe the inference model in multiSNV.
| Parameter | Description | Comments |
|---|---|---|
| n | Number of tumour samples | User-specified |
| SN | Allelic composition in normal sample | Estimated (one to two alleles allowed) |
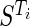 |
Allelic composition in tumour sample i | Estimated (one to three alleles allowed) |
| k | Denotes sample k | k ∈ {N, T1…Tn} |
| Mk | Number of alleles in allelic composition Sk | Computed from inferred Sk |
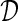 |
Pileup of bases and corresponding base qualities in all samples | Read from pileup file |
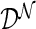 |
Pileup of bases and corresponding base qualities in normal sample | Read from pileup file |
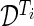 |
Pileup of bases and corresponding base qualities in tumour sample i | Read from pileup file |
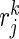 |
Allele supported by read j in sample k | Read from pileup file |
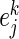 |
Error probability of read j in sample k | Read from pileup file |
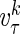 |
Total number of reads in sample k that support allele τ | Read from pileup file |
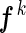 |
The probability distribution of alleles in sample k | Estimated by maximizing 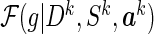
|
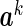 |
Vector of shape parameters of Dirichlet prior on 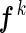
|
Uniform prior |
| μ | Mutation rate | User-specified (default is 3 × 10−7) |
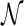 |
The set of values of SN with nonzero prior probability | All monoallelic and diallelic compositions (total of 10) |
| ϕz | Sampling probability of allelic composition 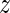
|
Integrated out |
| δz | Parameters of Dirichlet prior on ϕz of 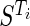
|
As described in Materials and Methods |
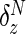 |
Parameters of Dirichlet prior on ϕz of SN | As described in Materials and Methods |
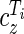 |
Number of tumour samples excluding Ti with allelic composition 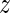
|
From most recent draw of the Gibbs sampler |
| nz | Number of tumour samples with allelic composition 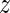
|
From most recent draw of the Gibbs sampler |
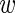 |
Scales pseudocounts for Dirichlet prior of allelic compositions | 10 × (n + 1) |
multiSNV analyses each location in the genome independently, so these parameters refer to a single genomic locus.
