Figure 4.
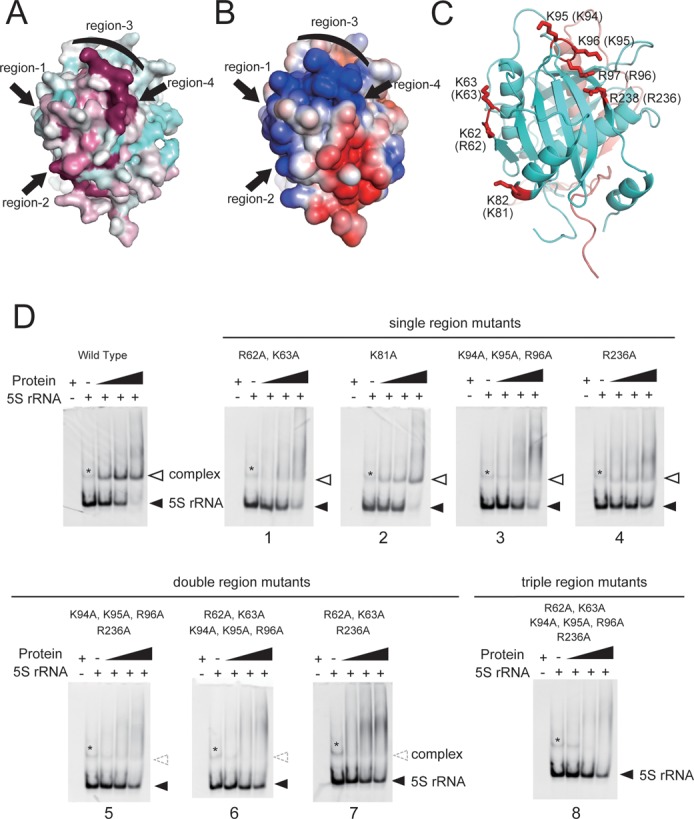
Evolutionarily conserved and electrically positive regions of ScRpf2 (region-1 to 4) were selected for mutagenesis experiments to test the significance of the interactions with 5S rRNA. (A) Sequence conservation is mapped onto the surface along with variable (cyan) and conserved (purple) residues using Consurf (28). (B) Electrostatic surface potential diagrams with positive (blue) and negative (red) electrostatic potentials are mapped onto a van der Waals surface diagram of the conserved surface patch using APBS (29) The color scale ranges between −3 kBT (red) and +3 kBT (blue), where kB is Boltzmann's constant and T is temperature. (C) Ribbon diagram of the Rpf2-Rrs1 core complex in the same orientation as in A and B. Four regions containing seven residues (red) were selected for mutation analysis. Residue numbers of ScRpf2 are shown. Letters in parentheses correspond to the residue numbers for AnRpf2. (D) Results of the gel shift assay. Sc5S rRNA (50 pmol) was incubated without factor or with 50, 100, or 200 pmol of ScRpf2-Rrs1 complex mutated variants; the denoted numbers correspond to the mutant No. (Supplementary Table S1). Wild type indicates ScRpf2ΔC-Rrs1ΔC purified using the same method used to purify other point-mutated variants. Asterisk (*) indicates the 5S rRNA dimer, as confirmed by a gel-filtration analysis and urea-PAGE. Results are shown as an ethidium bromide-stained gel.
