Figure 3.
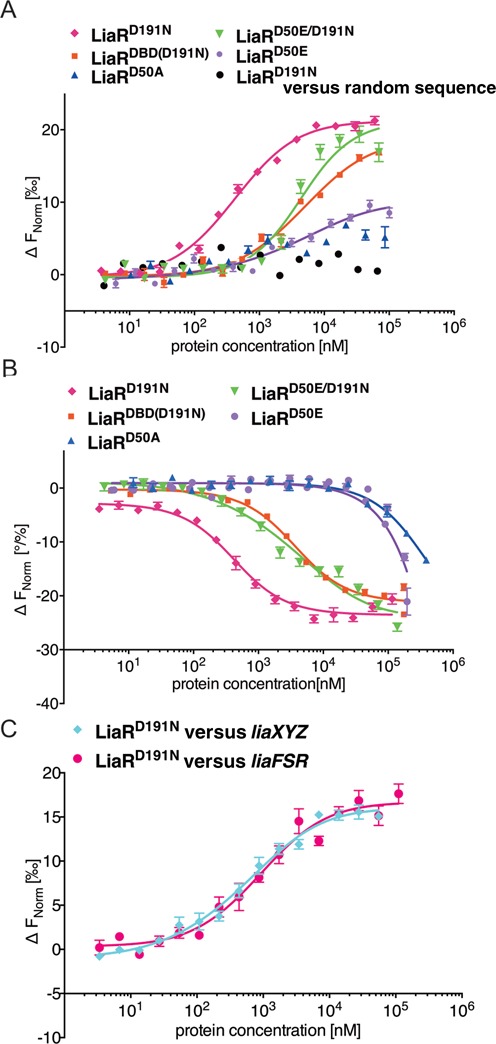
Adaptive mutant LiaRD191N that confers increased daptomycin resistance in E. faecalis S613 dramatically increases LiaR affinity for target DNA sequences. E. faecalis response regulator LiaR–DNA interactions were measured with MST. To determine the Kd, increasing concentrations of LiaRD191N was added to 40 nM of fluorescently labeled DNAs (Supplementary Information). Fnorm (normalized fluorescence) was plotted on the y-axis in per mil [‰] unit (meaning every 1000) against the total concentration of the titrated partner on a log10 scale on the x-axis (21). The resulting Kd values based on average from six independent MST measurements. Note, when the markers were increased in size for readability the error bars became covered in some cases. (A) The binding of LiaRD191N (magenta diamonds), LiaRD50E (purple circles), LiaRD50A (blue triangles), LiaRD50E/D191N (green triangles) and LiaRDBD(D191N) (red squares) to the consensus sequence within the liaXYZ operon or LiaRD191N with a random sequence (black circles). (B) The binding of LiaRD191N (magenta diamonds), LiaRD50E (purple circles), LiaRD50A (blue triangles), LiaRDBD(D191N) (red squares), LiaRD50E/D191N (green triangles) to the extended site of the liaFSR operon. (C) The binding of LiaRD191N to the entire contiguous protected regions (Figure 2B, D) of liaXYZ (cyan diamonds) and liaFSR (magenta circles).
