Figure 9.
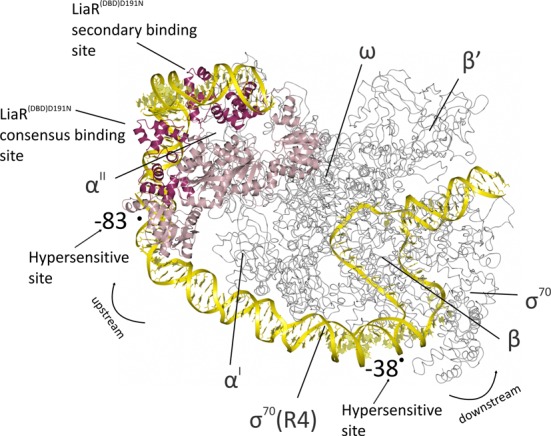
Model for the binding of the activated LiaR tetramer onto the regulatory sequences responsible for the LiaR-mediated cell envelope stress response. The LiaRD191N DNA binding domains bound to DNA sequences derived from the liaXYZ consensus and secondary sites (dark red; PDB: 4WUL and 4WU4) are modeled as an active tetramer using the protein alone tetramer structure of the S. aureus VraR receiver domains (pink; PDB: 4IF4). The structure of the E. coli RNA polymerase complex initiation complex (gray; PDB: 3IYD) was used to model the position of RNA polymerase and DNA. The combined model for the LiaR:DNA and RNA polymerase:DNA complexes (yellow) suggests a strong bend that is consistent with the crystal structures and DNaseI hypersensitive sites from our protection studies (arrows). Note that the RNA polymerase α’ and ω subunits are well poised for potential interactions with the LiaR tetramer.
