Figure 4.
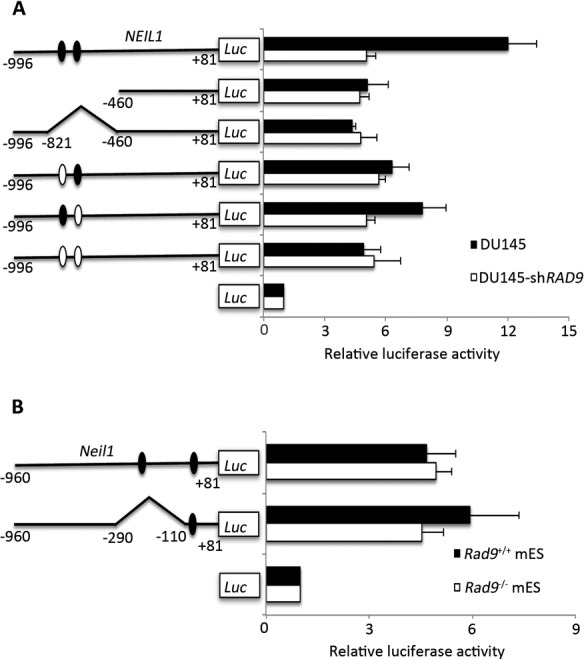
NEIL1 promoter-luciferase reporter activity in DU145 and mES cells with inherent or reduced levels of RAD9. Chimeric constructs of the NEIL1 promoter-luciferase reporter are schematically represented on the Y-axis. The X-axis indicates luciferase activity as fold above values obtained for the promoterless vector, pGL-Basic. (A) Human NEIL1 promoter sequence. DU145 (dark bar), DU145-shRAD9 (light bar) host cells. (B) Mouse Neil1 promoter sequences. mES Rad9+/+(dark bar), Rad9−/− (light bar) host cells. Error bars represent the standard deviation of three independent experiments. Luc, luciferase. Numbers on constructs in Y-axis correspond to nucleotide positions in promoters relative to the start of transcription. Dark ovals represent intact p53-binding sites; light ovals indicate mutation sites. Pointed regions of promoters contain RAD9 binding sequences, as per the Chip-qPCR data, and are deleted.
