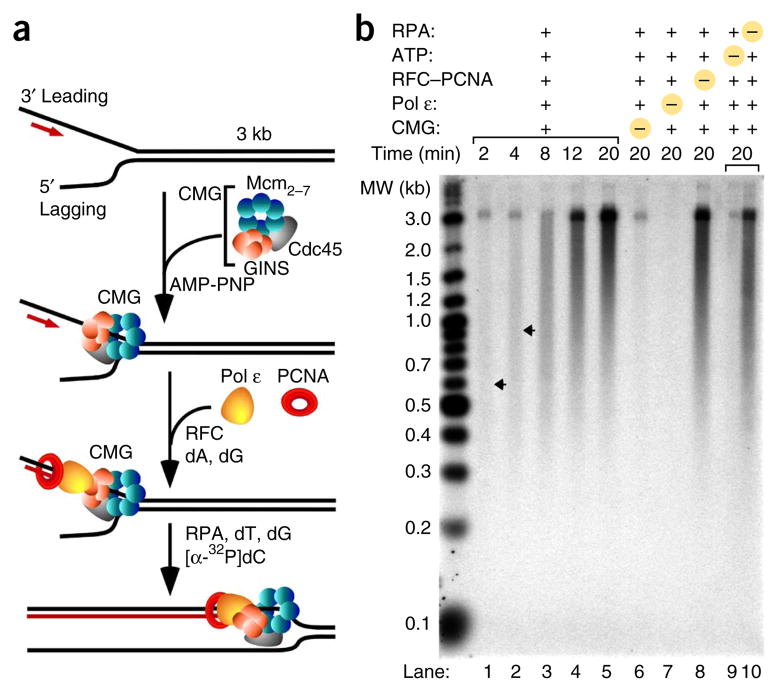
Figure 1.
Reconstitution of a leading-strand replisome with CMG, Pol ε, RFC, PCNA and RPA. (a) Scheme of the assay. The 3.2-kb forked DNA substrate was constructed by ligation of a synthetic primed replication fork to a 3.2-kb duplex created with nucleotide bias to enable specific labeling of the leading strand with [32P]dCTP. CMG was assembled on the DNA with AMP-PNP in a 10-min preincubation; then Pol ε, PCNA and RFC were added for 2 min along with cold dATP and dGTP before replication was initiated upon addition of RPA, ATP, dTTP and [32P]dCTP. Experimental details and protein concentrations are in Online Methods. (b) Alkaline agarose gel of reaction products. Lanes 1–5, time course of the full reaction; lanes 6–9, reactions with indicated components omitted. Arrows indicate the peak lengths of DNA product at 2 and 4 min. Quantitation of the results is in Supplementary Figure 2. MW, molecular weight.