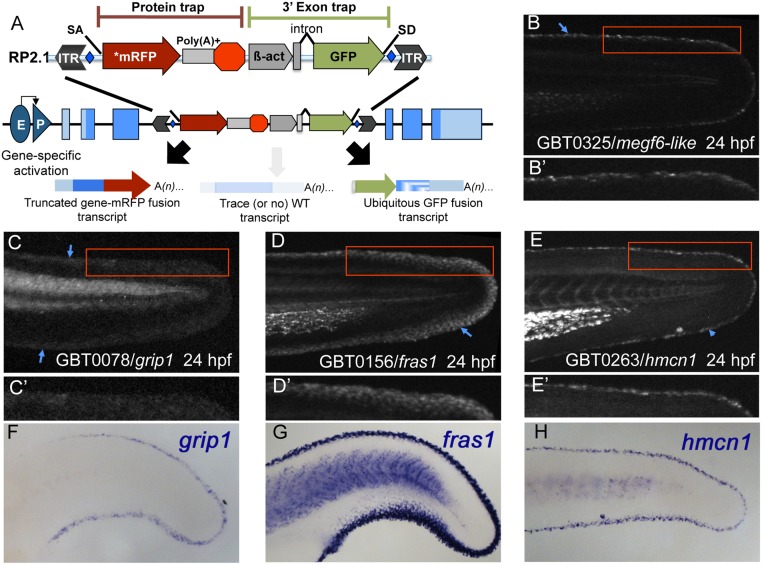
Fig 1. Gene-break transposon—based protein trapping identifies known and new epidermal median fin fold loci.
(A) A schematic of the RP2.1 gene-break transposon (GBT) vector used in this study. Gene-breaking activity occurs when an endogenous locus with a GBT insertion is transcribed. The vector-supplied splice acceptor (SA) in the 5’ protein trap cassette intercepts the endogenous splicing machinery and transcript, redirecting them to read directly into an AUG-free mRFP sequence (*mRFP). That event generates a fusion transcript by tagging the 5’ portion of the endogenous transcript with mRFP. When translated, the mRFP fusion transcript will produce a potentially mutagenic truncated protein product. Simultaneously, the 3’ exon trap cassette uses the vector-supplied splice donor (SD) to create a GFP fusion transcript with the remaining downstream endogenous transcript. GBT alleles are revertible because loxP sites (blue diamonds) flank the cassettes, allowing the mutagenic elements to be excised in the presence of Cre recombinase. (B-E’) At 24 hours post-fertilization (24 hpf), GBT-generated mRFP fusion proteins from megf6a mn0325Gt (B), grip1 mn0078Gt (C), fras1 mn0156Gt (D), and hmcn1 mn0263Gt (E) localize along MFF edges. (B’, E’) Both megf6a mn0325Gt (B’) and hmcn1 mn0263Gt (E’) localize within a narrow region along the MFF edge (blue arrowheads). (C’, D’) grip1 mn0078Gt (C’) and fras1 mn0156Gt (D’) localization also follows the fin fold edge (blue arrowheads), though they are distributed somewhat more diffusely than are megf6a mn0325Gt (B’) and hmcn1 mn0263Gt (E’). (F-H) Whole-mount in situ hybridization (WISH) in 24 hpf wild-type embryos reveals similar MFF expression patterns of endogenous grip1 (F), fras1 (G), and hmcn1 (H) genes. The mRFP fusion protein localization patterns observed in the respective GBT lines recapitulate endogenous gene expression (C, D, E).