Abstract
Temozolomide (TMZ) increases the overall survival of patients with glioblastoma (GBM), but its role in the clinical management of diffuse low-grade gliomas (LGG) is still being defined. DNA hypermethylation of the O6-methylguanine-DNA methyltransferase (MGMT) promoter is associated with an improved response to TMZ treatment, while inactivation of the DNA mismatch repair (MMR) pathway is associated with therapeutic resistance and TMZ-induced mutagenesis. We previously demonstrated that TMZ treatment of LGG induces driver mutations in the RB and AKT-mTOR pathways, which may drive malignant progression to secondary GBM. To better understand the mechanisms underlying TMZ-induced mutagenesis and malignant progression, we explored the evolution of MGMT methylation and genetic alterations affecting MMR genes in a cohort of 34 treatment naïve LGGs and their recurrences. Recurrences with TMZ-associated hypermutation had increased MGMT methylation compared to their untreated initial tumors and higher overall MGMT methylation compared to TMZ-treated non-hypermutated recurrences. A TMZ-associated mutation in one or more MMR genes was observed in five out of six TMZ treated, hypermutated recurrences. In two cases, pre-existing heterozygous deletions encompassing MGMT, or an MMR gene, were followed by TMZ-associated mutations in one of the genes of interest. These results suggest that tumor cells with methylated MGMT may undergo positive selection during TMZ treatment in the context of MMR deficiency.
Keywords: Low-grade glioma, Temozolomide, Hypermutator, Mismatch Repair, MGMT
Introduction
Diffuse LGG are infiltrative brain tumors which include World Health Organization (WHO) grade II astrocytomas, oligodendrogliomas, and oligoastrocytomas [33]. Surgical resection is the primary therapeutic intervention, though LGG recur and may undergo malignant progression to a higher histological grade including grade IV secondary GBM. Therefore, in patients with clinical risk factors [42, 51], postoperative adjuvant treatment is often utilized. The addition of TMZ to postoperative radiotherapy prolongs progression free survival (PFS) and overall survival (OS) in high-risk LGG patients, and chemotherapy instead of irradiation might be as effective [3, 46] and defers the risk of late radiation-induced cognitive deterioration [14]. Moreover, postoperative TMZ or irradiation of LGG has been associated with improved quality of life, better seizure control, and longer progression-free survival [6, 30, 38, 43, 49].
TMZ is an alkylating agent that methylates the O6 position of guanine. The DNA repair protein MGMT removes O6-methyl groups induced by TMZ. Initial studies assaying DNA methylation in the MGMT gene body rather than promoter showed a direct correlation between MGMT methylation and expression [19, 41]. When the MGMT promoter is hypermethylated however, MGMT expression is decreased and TMZ-induced DNA damage persists [12, 13, 27]. O6-methylguanine pairs with thymine instead of cytosine during DNA replication. MMR can recognize and repair these mismatches through MutS and MutL complexes. MSH2 and MSH6 form the MutSα complex, which identifies base-base mismatches and small insertion-deletion-loops (IDLs). MSH2 and MSH3 form the MutSβ complex which identifies large IDLs. MutS complexes directly with MutL, an MLH1/PMS2 dimer, to the site of DNA damage [20;21]. Removal of the thymine that is base paired with O6-methylguanine is followed by repair synthesis that reinserts thymine, leading to repeated attempts to repair the same base. This futile cycling of repair has been linked to DNA double strand breaks and apoptosis, the apparent mechanism of TMZ-induced cytotoxicity [16].
Inactivation of the MMR pathway is a mechanism of resistance to TMZ in primary GBMs and also leads to TMZinduced mutagenesis [8, 18, 26, 63]. In MMR deficient cells, the base pairing of O6-methylguanine with thymine persists, and upon DNA replication results in nucleotide transitions from guanine to adenine. TMZ-associated hypermutation has been observed in GBM [9–12], in cells treated with TMZ in vitro [8] and in unpaired posttreatment tissue samples [1, 5, 15, 16, 26]. In contrast to MMR, the impact of MGMT activity on the relative amount of cytotoxicity versus mutagenicity is much less clear. Furthermore, while MGMT methylation is associated with longer overall survival in GBM patients treated with TMZ [25], it is unclear whether this biomarker has the same prognostic value in patients with IDH1 mutated LGG [17, 54, 59].
We recently identified hypermutation in a subset of TMZ-treated recurrent GBMs that arose from IDH1-mutant astrocytic LGG [29]. Post-TMZ recurrences had a 39–133 fold increase in the mutation rate relative to their treatment naïve initial LGG, more than 98% of which C>T/G>A mutations which are associated with TMZ-induced mutagenesis (Supplementary Table S1) [5]. TMZ-associated mutations resulted in deregulation of RB mediated cell cycle control and hyperactivated AKT-mTOR signaling, suggesting TMZ-induced hypermutation may drive malignant progression. However, it is unclear why hypermutation developed in only six of the ten LGG treated with TMZ. To better understand the mechanism of hypermutation, here we examined the stepwise development of DNA repair deficiency and subsequent TMZ-associated hypermutation using a cohort of 34 initial LGG and their patient-matched recurrence, including 23 pairs for which exome sequencing data was available. Because TMZ-induced hypermutation in LGG was associated exclusively with GBM recurrence, this study is important to understanding and ultimately avoiding TMZ-associated hypermutation and malignant progression.
Methods
Sample acquisition
Patient inclusion in this cohort was dependent upon 1) an initial diagnosis of WHO grade II diffuse astrocytoma, oligodendroglioma, or oligoastrocytoma; 2) available tumor tissue from an initial tumor and a subsequent recurrence; 3) information on post-surgical treatment. A majority of the samples have been used in previous studies (Supplementary Table S2)[29, 56]. Tumor samples were fresh-frozen or formalin fixed paraffin embedded (FFPE) tissues. Sample use was approved by the Committee on Human Research at UCSF; the Ethics Committee of the University of Tokyo; and the Medical Ethics Committees of the Dutch hospitals VU University Medical Center Amsterdam, Radboud University Medical Center Nijmegen, Isala Klinieken Zwolle and Erasmus Medical Center Rotterdam, and the Linköping University Hospital, Sweden.
DNA isolation
Genomic DNA from tumor and normal tissue samples of patients 01–38 was extracted with either the QIAGEN FFPE DNA extraction kit (Qiagen, Valencia, CA) following the manufacturer’s instructions or isolated by a standard phenol chloroform extraction as previously described [29]. FFPE blocks of initial tumor and recurrences of patients 90–302 were cut into sections of 3–5 µm thickness for pathological evaluation on hematoxylin and eosin stained slides. For each sample, an area was delineated that contained >60% tumor cells. The corresponding area on subsequent sections of 10 µm was used for DNA isolation extracted with the QIAGEN FFPE DNA extraction kit[56].
Bisulfite treatment, PCR, cloning and sequencing of MGMT
MGMT methylation status was assessed for all patients (Fig. 1a and Fig. S1) [24]. DNA (>100ng) was bisulfite-treated for 2.5 hours with the EZ DNA methylation Gold kit (Zymo Research, Irvine, California) according to the manufacturer’s instructions. Bisulfite-converted DNA was amplified by PCR using the following primers corresponding to the MGMT promoter: forward GGATATGTTGGGATAGTT and reverse ATCGTTAATAAGTCAAGCTC. Gel extraction of the amplified DNA was performed with the QIAEXII gel extraction Kit (Qiagen, Germantown, Maryland). Four to six microliters of PCR product was cloned using a pCR2.1/TOPO TA sequencing kit (Invitrogen, Carlsbad, California). Individual bacterial clones were subjected to PCR using vector-specific primers and a minimum of 9 individual PCR clones were sequenced per tumor sample. Bisulfite sequence data of the MGMT promoter were analyzed with BISMA [27, 48]. The bisulfite conversion rate was monitored in all reactions at non-CpG cytosines, which are typically unmethylated and converted. For comparison, the methylation status of the MGMT promoter in bisulfite-treated DNA was also determined in a subset of the samples by standard, non-quantitative methylation-specific PCR (MSP) [16].
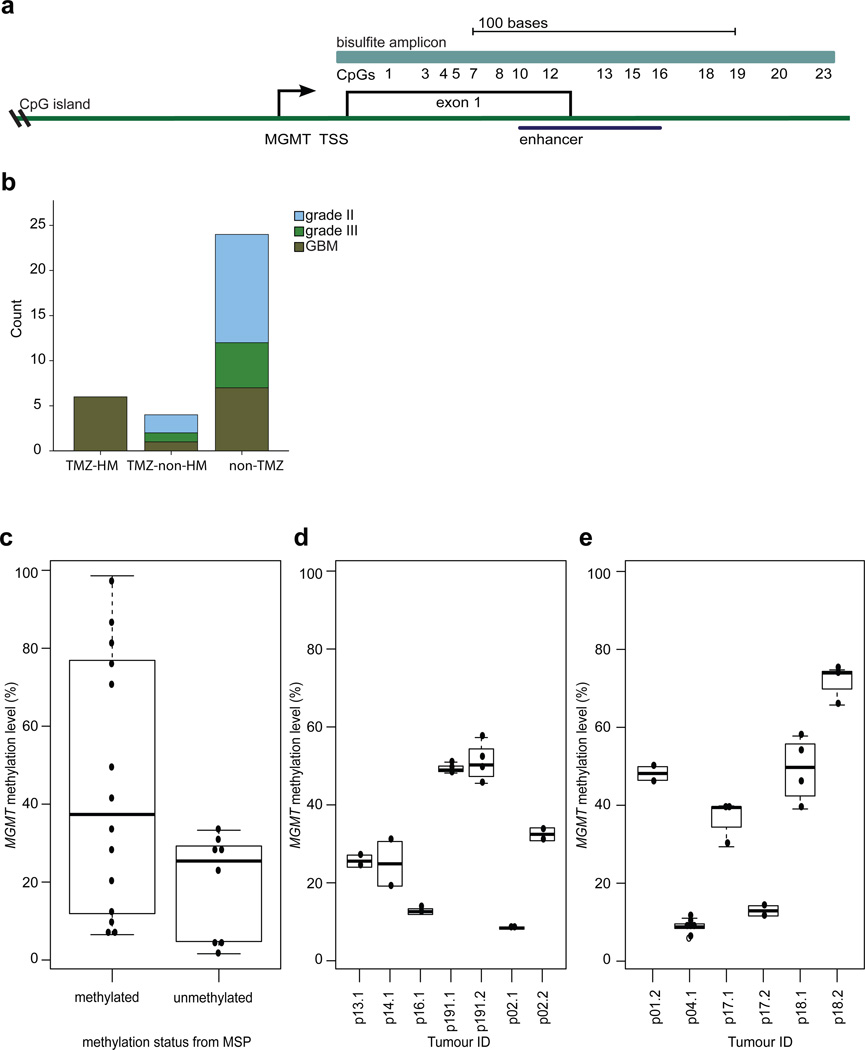
Fig. 1.
Comparison of MGMT methylation in different assays and across tumor regions. a Position of the MGMT bisulfite amplicon (light blue) encompassing the 23 CpGs assessed in this study, the position of the MGMT CpG island (green), and the enhancer region encompassing CpGs 10–16 (dark blue). TSS= transcription start site. b Distribution of histological subtypes and grades of the recurrent tumors in the three groups. c Comparison of the binary outcome of MSP (x-axis) to MGMT methylation level of CpGs 10 to 16 determined by bisulfite, PCR and sequencing of 10 or more independent clones (y-axis). d The degree of variation in methylation levels was determined in replicate experiments from independent aliquots of the same genomic DNA isolation (y-axis) with bisulfite and sequencing of the MGMT promoter in individual samples (x-axis). e MGMT methylation levels (y-axis) in spatially distinct regions of individual tumors (x-axis). Sample designations are the patient (p) number followed by .1 for initial tumor and .2 for the recurrent tumor.
Identification of somatic mutations and copy number aberrations in MGMT and MMR genes
The identification of MMR pathway alterations was limited to those for which sufficient tumor DNA and matched normal DNA was available for exome sequencing. The mutational and copy number status of MGMT as well as the key MMR pathway genes MLH1, MLH3, MSH2, MSH3, MSH5, MSH6, PMS1, and PMS2 [28] were assessed from the exome sequencing data. For patients 01–24, somatic mutations and copy number aberrations in genes of interest were identified as previously described [29]. Nine new exome sequencing datasets were also generated for this study using the Agilent SureSelect Target Enrichment System Protocol (Version 1.0 September 2009) with the SureSelect Human All Exon 50Mb kit (Agilent Technologies) according to the manufacturer's instructions. Paired-end reads of 76bp or 100bp in length were generated from Illumina HiSeq 2000 or 2500 instrumentation. Paired-end sequencing data from exome capture libraries were aligned to the reference human genome (build hg19) with the Burrows-Wheeler Aligner (BWA) 0.5.10 [32]. Single-nucleotide variants (SNVs) were detected with MuTect [11], and indels were detected with Pindel [62], followed by custom filters to remove false positives [29]. All candidate mutations were subsequently validated with PCR amplification of the target region from tumor and matched normal genomic DNA followed by conventional Sanger sequencing. Copy number segmentation was performed with an adaptation of circular binary segmentation (CBS) [29, 57]. We identified germline heterozygous SNPs from the matched normal exome of each patient tumor using the UnifiedGenotyper [35]. From only those SNPs present in dbSNP (Build ID: 132) (http://www.ncbi.nlm.nih.gov/SNP/) and with a coverage level of 10 or more reads, we calculated their minor allele frequency in all exomes of each patient (initial tumor, recurrence, and patient-matched normal) and used these to infer genomic regions with loss of heterozygosity (LOH). Regions of LOH were then correlated with DNA copy-number alterations. For patients 171–296 copy number aberrations at genes of interest were identified from low-coverage whole genome sequencing as previously described [50, 56]. Copy number segmentation was performed by CBS [29] and gains and losses were identified using CGHcall [55].
Significance tests
For differences in methylation, the initial comparison between subgroups was done using the Kruskal-Wallis test (the nonparametric alternative to ANOVA), followed by subsequent post hoc testing using the Wilcoxon rank-sum or signed-rank test (on data from tumor pairs) for two group analyses. P values below 0.05 were considered statistically significant.
Results
Histological features and disease course of the cohort
Clinical and genomic data for many of the paired initial and recurrent tumor samples used in this study have been described previously [29, 56]. In total, we studied 87 samples from 34 LGG patients. Samples from spatially distinct regions of the tumor were available for six surgeries. Ten of the 34 patients received adjuvant TMZ. Accordingly, we divided the cohort into three groups based on the clinical and exome sequencing data; patients with recurrent tumors displaying a TMZ-associated hypermutator phenotype (TMZ-HM, n=6), TMZ-treated patients without a TMZ-associated hypermutator phenotype (TMZ-non-HM, n=4) and patients not treated with TMZ (non-TMZ, n=24) (Supplementary Table S2). While the overall size of the cohort is relatively modest, very few studies have reported genomic and epigenomic evolution in similarly sized cohorts of paired initial LGG and recurrent tumors [58]. In the TMZ-HM group, all six tumors recurred with GBM histology. Relative to the TMZ-HM group, the number of recurrent tumors with GBM histology was variable in the TMZ-non-HM group and non-TMZ group, (ANOVA, p-value 0.013) (Fig. 1b).
MGMT methylation level is similar across assays, technical replicates and spatially distinct samples
Methyl specific-PCR (MSP) is used in clinical tests for MGMT methylation status and yields a low-resolution, non-quantitative binary call of methylated or unmethylated. MGMT methylation levels in tumors span a full range from unmethylated to fully methylated at each CpG however. We therefore compared MSP to a more quantitative, single-CpG resolution method involving bisulfite treatment, PCR, cloning and sequencing in 18 samples. The genomic region assessed by the bisulfite sequencing approach assesses methylation level at each of 23 CpG sites in the MGMT promoter and enhancer, including the region covered by the MSP assay which spans CpG sites 10 to 16 [2, 34], a previously reported differentially methylated region 2 (DMR2) covered by CpGs 3 to 20 [34], and CpG site 13 which has prognostic value in GBMs [2] (Fig. 1a). In eight samples classified as MGMT-unmethylated by MSP, median methylation level was 25.4% (range 1.6–28.6%), and in 10 samples that were methylated according to MSP, median methylation level was 37.4% (range 6.5–98.6%) (Fig. 1c). To test the reproducibility of bisulfite sequencing approach, experiments of seven samples were repeated on independent aliquots from the same genomic DNA isolation. Very little variation in methylation level was observed between replicate experiments (Fig. 1d). An analysis of spatially distinct regions in samples obtained from the same surgery revealed that there was also very little variation in MGMT methylation levels within a tumor at a given time point (Fig. 1e). This limited intratumoral heterogeneity of MGMT methylation level provided evidence that the result of a single sample was likely to be representative for LGG, as previously shown in GBM [15, 21, 23].
Increase in MGMT methylation level associated with temozolomide-induced hypermutation
MGMT methylation levels varied widely between patients. Across the whole cohort, the median methylation level of initial tumors was 29.7% (range 6.1–70.9%) and the median methylation level of recurrent tumors was also 29.7% (range 3.9–79.1%), (p value 0.49) (Supplementary Material). Between the three subgroups, median methylation levels in initial tumors were not significantly different (TMZ-HM 38.3% vs. TMZ-non-HM 17.8%, p value 0.15; TMZ-HM vs. non-TMZ 33%, p value 0.33), while overall methylation level in recurrent tumors of the TMZ-HM group (median 55%) was significantly higher compared to recurrent tumors of the TMZ-non-HM (median 20%) and non-TMZ (median 28%) groups (p value 0.013 and 0.03 respectively) (Fig. 2). Patient 24 was not included in this analysis, as methylation data were confounded by low tumor purity in the initial and first recurrent tumor, complicating the interpretation.
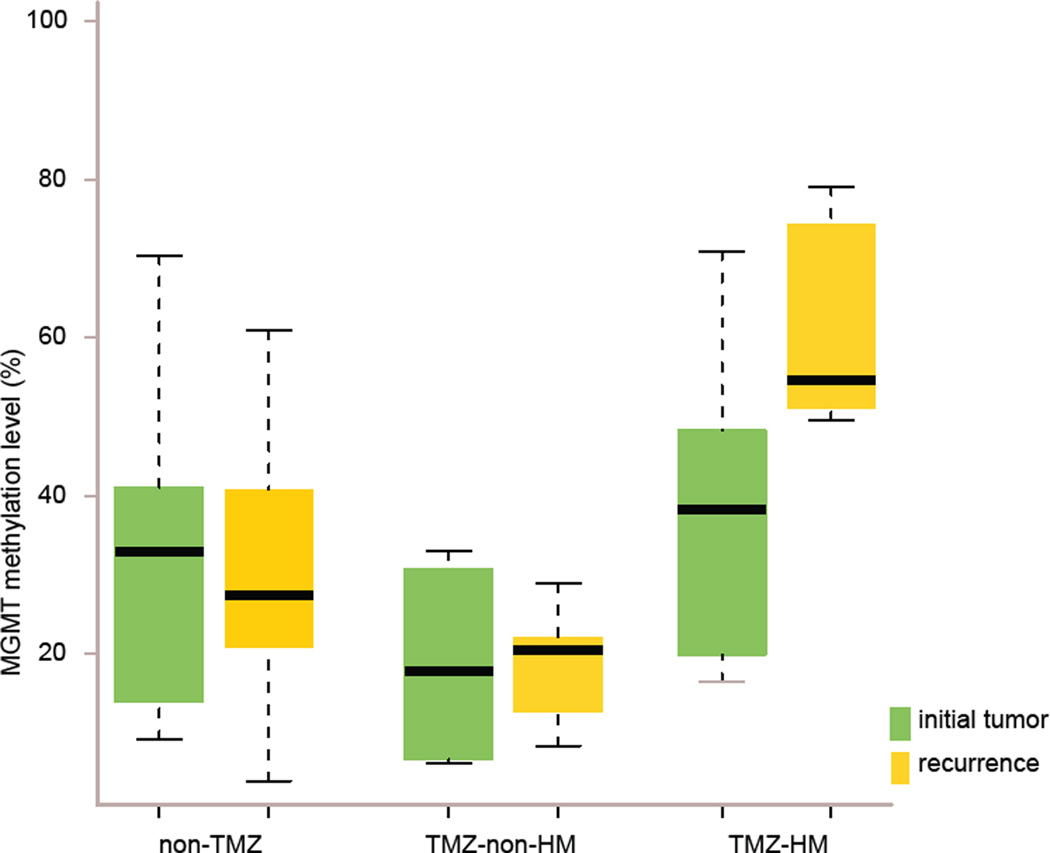
Fig. 2.
Significantly elevated MGMT methylation in TMZ-associated hypermutated recurrent tumors that arise from LGG. MGMT methylation levels in initial (green) and recurrent (yellow) tumors of three patient subgroups; non-TMZ= patients not treated with TMZ, TMZ-non-HM= patients treated with TMZ without a hypermutated recurrent tumor, TMZ-HM= patients treated with TMZ with a hypermutated recurrent tumor.
We explored how methylation levels changed over time by performing paired analysis in initial and recurrent tumors of the three subgroups TMZ-HM, TMZ-non-HM, non-TMZ. The change in methylation level from initial to recurrence in the TMZ-HM group was non-significant but showed a trend (p value 0.063). This is supported by the consistent increase in methylation level in this subgroup. This pattern was significantly different from the variable patterns of change over time in the TMZ-non-HM and non-TMZ groups (TMZ-HM vs. TMZ-non-HM p value 0.050; TMZ-HM vs. non-TMZ p value 0.005) (Fig. 2). Eleven of 23 individual CpGs were significantly more methylated in the recurrent tumors of the subgroups TMZ-HM vs. TMZ-non-HM (CpGs 1–6, 8–10, 12 and 13, p values 0.012–0.044).
Evolution of DNA repair deficiency in the TMZ-HM group
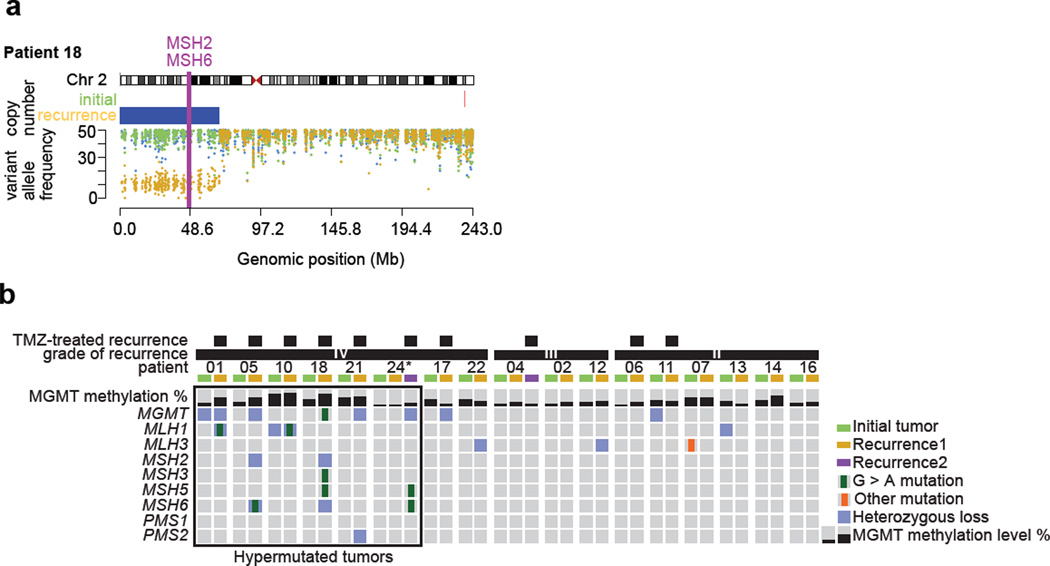
In the initial tumors of the TMZ-HM group (n=6) no MMR gene mutations were detected, however DNA repair may have been impaired by the heterozygous loss of MGMT in patient 01 and MLH1 in patient 10. In contrast, five of the six TMZ-HM GBMs contained a TMZ-associated mutation in one of the MMR genes, concurrent with deletion of the other allele or deletion encompassing another MMR gene or MGMT (Fig. 3a and b, Fig. S2). We also identified a clonal TMZ-associated mutation in MGMT of unknown significance in the recurrent tumor of patient 18. In the initial and recurrent tumors of the TMZ-non-HM (n=4) patients, MMR pathway genes were intact, but heterozygous loss of MGMT was detected in the initial tumor of patient 11 and the recurrent tumor of patient 17. Interestingly, the recurrent tumor of patient 11 grew out from an earlier cell that retained both alleles of MGMT, while the recurrent tumor of patient 17 had decreased levels of DNA methylation at MGMT (initial 28.3%, recurrence 12.7%), indicating that in both cases MGMT levels may not have been impaired during TMZ treatment and recurrence. In the non-TMZ group (n=24), mutational analysis was performed in patients of which sufficient DNA from matched normal and initial and recurrent tumor was available (n=7). In these seven cases only one MMR mutation was detected, an MSH3 mutation in the initial tumor but not in the recurrent tumor of patient 07. DNA copy number status of MGMT and MMR genes was available for 21 of 24 patients in the non-TMZ subgroup. Genomic loss affecting the MGMT region was detected in five initial and seven recurrent tumors of the non-TMZ subgroup. Similarly, deletion encompassing an MMR gene was shared between the initial and recurrent tumors of five cases, while four patients acquired a deletion encompassing a MMR gene at recurrence (Supplementary Table S3). As these patients did not receive TMZ, it is not known how TMZ treatment may have affected MGMT methylation levels at recurrence, or if the identified genetic alterations to MGMT and MMR genes may indicate a susceptibility to hypermutation.
Fig. 3.
Sequential acquisition of DNA repair deficiency exclusively in LGG patients that were treated with TMZ and had a hypermutated recurrence. a Exome derived copy number status and germ-line variant allele frequency across chromosome 2 encompassing MSH2 and MSH6 in the initial and recurrent tumor of patient 18. b Methylation level of MGMT, mutations and copy number status of MGMT and MMR-related genes in initial LGG and paired recurrent tumors. The panel includes patients for whom exome sequencing data was available, * indicates samples with a proportion of tumor cells lower than 50%.
Discussion
We compared spontaneous and treatment-associated evolution of DNA repair deficiency in a cohort of 34 initial LGG and their patient-matched recurrences. Our data suggest MGMT and MMR mediated DNA repair may be compromised by sequential and coincident loss of heterozygosity, methylation [45], and TMZ-associated mutation, although repair activity could not be tested directly. Considering prior studies of TMZ-treated GBM patients [1, 8, 26, 63] and cells treated with TMZ in vitro [5], our results suggest that TMZ-induced hypermutation is the consequence of a TMZ resistance mechanism in LGG. This putative mechanism is not fully understood, but may be induced directly by the mutagenic action of TMZ on DNA repair genes, in combination with pre-existing and concurrent copy number alterations in cells with a higher level of MGMT methylation. The resistance mechanism appears to involve a switch from toxicity to tolerance of TMZ-induced DNA damage. The sequential acquisition of genetic and epigenetic change in MGMT and MMR genes in the TMZ-HM group differs notably from the patterns in patients that did not receive TMZ, and in TMZ-treated but not hypermutated patients.
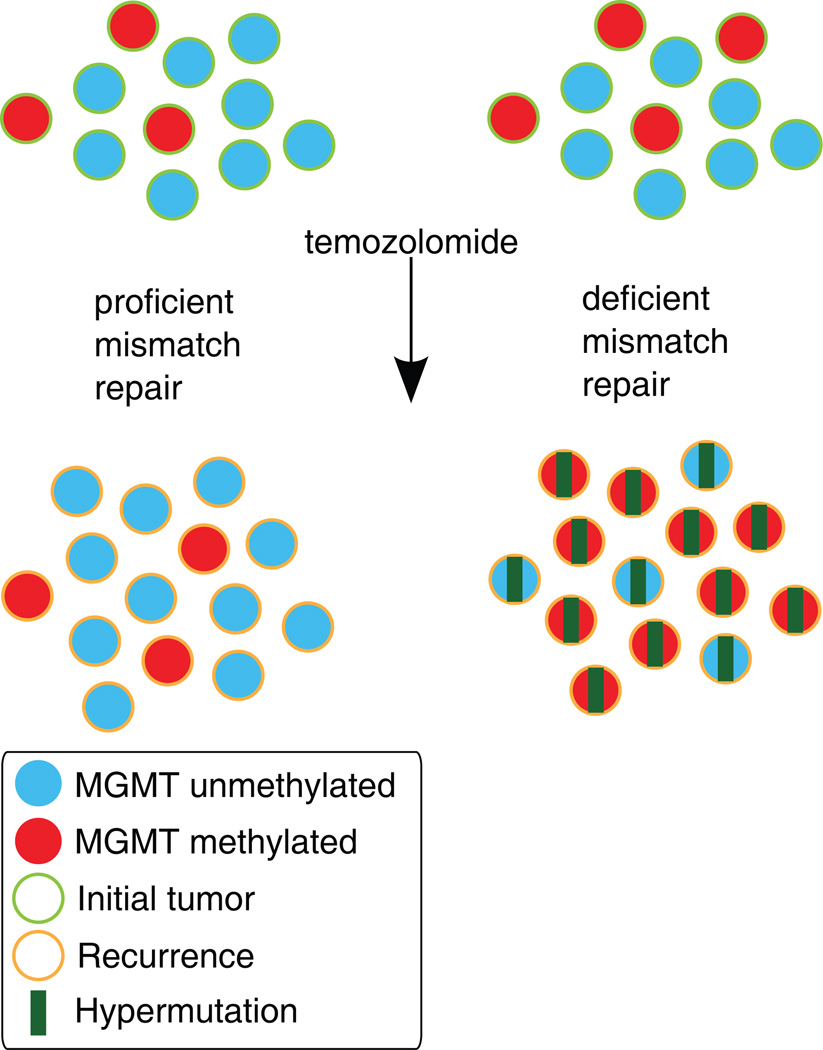
We observed a consistent increase over time in MGMT methylation level, which was not detected in LGG patients without a TMZ-associated hypermutator phenotype. The apparent positive selection of MGMT hypermethylated cells and a corresponding decrease in MGMT expression may predispose a cell to persistent 0–6-methylguanine lesions and acquisition of MMR gene mutation, enabling hypermutation from subsequent rounds of TMZ treatment (Fig. 4). MGMT activity also may be decreased by TMZ treatment itself, as the MGMT protein is not regenerated following repair [53].
Fig. 4.
A working model of the effect of an impaired MMR system on clonal outgrowth of MGMT methylated cells during acquisition of the TMZ-associated hypermutator phenotype and TMZ-associated malignant progression. Left: When MMR is intact, TMZ treatment induces cell death in MGMT methylated cells. The histology of the recurrent tumor is variable. Right: When MMR is deficient, TMZ treatment fails to induce cell death and MGMT methylated tumor cells may expand, become hypermutated, and undergo malignant progression to GBM.
Other studies have addressed temporal changes of MGMT methylation in smaller cohorts of grade II astrocytomas [22, 31, 36] and GBMs [7, 9, 39] with MSP only, and without mutational and copy number analysis. In the present study, bisulfite sequencing of the MGMT promoter in 34 paired initial and recurrent tumors enabled detailed, quantitative analysis of temporal evolution in individual patients. Given the sample size, a meaningful comparison of MGMT methylation change was not possible for GBM recurrences that were HM (n=6) versus GBM recurrences in the TMZ-non-HM subgroup (n=1). We observed a distinct pattern of increased MGMT methylation level between initial and patient-matched, TMZ treated and hypermutated recurrences.
Exposure of cells to a mutagen such as TMZ will result in a different set of mutations in each cell within the population. However, our detection of somatic mutations in MMR-related genes in hypermutated DNA derived from bulk samples strongly suggests that these recurrences are derived from clonal expansion from a very small number of hypermutated cells [26]. The mutations are predominantly C>T/G>A transitions, the type of mutation known to be induced by TMZ treatment. Literature on the functionality of these mutations varies, for example somatic mutations MLH1 P648L and P640S in the TMZ-treated recurrent tumors of patients 01 and 10 also occur in the germline of families with hereditary nonpolyposis colon cancer, significantly affect MLH1 protein function, and are predicted to be pathogenic [10, 20, 44, 52]. The splice site mutation in MSH3 of patient 18 leads to a single nucleotide shift of the splice acceptor site, resulting in an out-of-frame transcript and premature truncation of the protein. However, the role of MSH3 in cancer is less clearly defined [40]. In MMR deficient cells, futile cycling of MMR repair does not occur, enabling this type of mutation. The C>T/G>A mutation also occurs spontaneously, however the extreme number of new mutations, the strong bias towards C>T/G>A versus other mutations, and the occurrence of hypermutation after TMZ treatment but not in patient-matched pre-treatment samples suggests TMZ is the predominant source. The proportion of tumors developing a hypermutation profile after TMZ treatment in our series is 60% (6 out of 10). This cohort and others [1, 26] are too small to determine the actual incidence of hypermutated diffuse gliomas after alkylating agent chemotherapy.
Biomarkers of susceptibility to TMZ-associated hypermutation could have significant clinical value. Rare germline and somatic MSH6 mutations that might affect how cells respond to TMZ have been detected in patients with untreated anaplastic oligodendrogliomas and GBMs [37, 47]. Within our small cohort, we found that loss of heterozygosity spanning MMR genes was unique to the TMZ-HM group relative to TMZ-non-HM group. In three TMZ-HM patients the initial tumor showed deletion of MGMT or an MMR gene. The copy number data of two of the other initial tumors from the TMZ-HM group was ambiguous. In a study of MMR protein expression assessed by immunohistochemistry decreased expression was observed for MLH1, MSH2 and MSH6 in a subset of initial low- and high-grade astrocytomas, but DNA copy number status and paired recurrences were not assessed [47]. A larger cohort of paired samples will be needed to determine if loss of heterozygosity of MGMT and/or MMR genes in initial tumors has predictive or prognostic value. Contrary to primary GBM, where copy number loss of the entire chromosome 10 is a frequent event [4], we observed variability in the size of the region lost in initial LGG and secondary GBM in our cohorts [56]. MGMT hypermethylation and corresponding impaired MGMT activity prior to TMZ treatment could also be a predisposing factor, but we did not detect statistically significant differences when analyzing MGMT methylation level alone between the initial tumors of the TMZ-HM and TMZ-non-HM subgroups. An alternative hypothesis is that, because TMZ-HM tumors appear to derive from a very limited number of cells, MGMT methylation in a small number of cells in the initial tumor may allow positive selection and hypermutation. Other studies with variable designs, and predominantly examining HGGs, were also unable to identify a correlation between MGMT methylation and MMR status [18, 37]. Similar to GBM [15, 21, 23], variation in MGMT methylation levels among multiple regions of the initial LGG of our patients was negligible, suggesting that single samples of the initial and recurrent tumor may be sufficient to elucidate temporal patterns. However, because the TMZ-HM group had an increased level of MGMT methylation relative to more variable patterns in the other groups, recurrences in the TMZ-HM group may exhibit greater intratumoral heterogeneity if the initial tumor resection was incomplete and sampling at recurrence included hypermutated and non-hypermutated regions.
The results presented here and in prior studies [1, 5], along with the well-established mechanisms of DNA repair by MMR and MGMT, further suggest that compromised DNA repair contributes to the onset of hypermutation and subsequent malignant transformation. Taken together, the data suggest a working model in which a hypermutated tumor arises through clonal expansion of cells with high levels of MGMT methylation, pre-existing loss of heterozygosity of a key MMR gene and/or MGMT, and TMZ-associated mutation in MMR genes. Tumor tissue and clinical data from LGG patients participating in clinical trials with TMZ treatment will be required to follow-up these initial findings [60, 61] and to assess the clinical relevance of the TMZ-associated hypermutator phenotype.
Supplementary Material
Acknowledgments
Disclosures and acknowledgements
This project was generously supported by Accelerated Brain Cancer Cure; The Grove Foundation; The TDC Foundation; The Anne and Jason Farber Foundation; and a generous gift from the Dabbiere family, UCSF Brain Tumor SPORE grant (NIH P50CA097257) ) (A.M., J.F.C., B.T., S.M.C., M.S.B.), the Dutch Cancer Society (KWF) grant number 2009–4470 and personal travel grant (H.F.v.T), foundation ‘STOPHersentumoren’, Edli foundation, LiU Cancer Research Network, The Medical Research Council of Southeast Sweden. Additional support by the National Institute Of General Medical Sciences T32GM008568 (T.M.), the National Institutes of Health 1T32CA15102201, the National Cancer Institute R01CA169316 (J.F.C.), Sontag Foundation (B.T.), NCI RO1 (R01 CA163687) (A.M.). This project was supported in part by a research program of the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct) (A.M., N.S., and H.A.), Grant-in-Aid for Scientific Research on Innovative Areas (No. 23134501) (A.M.), and Grant-in-Aid for Scientific Research (No. 24221011) (H.A.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We are grateful to our Dutch collaborators Guus Beute and Ruth Fleischeuer from St. Elisabeth hospital, Wimar van den Brink and Miek Havenith from Isala hospital, Hans Baayen, David Noske and Philip de Witt Hamer from VU University Medical Center and Anja M. Gijtenbeek from the Radboud University Medical Center Nijmegen for providing samples and clinical data. We thank collaborators from the South-East Sweden Brain Tumor Group for providing samples and clinical data; neurosurgeon Peter Milos, clinical oncologists Anna-Lotta Hallbeck, Linköping Charlotte Bratthäll, Kalmar and Michael Strandéus, Jönköping.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
Whole exome sequence data are uploaded to the European Genome-phenome Archive (EGA) for patients 1–23 (accession number EGAS00001000579), and shallow whole genome sequencing data of patients 9, 161–296 (EGAS00001000643). Data of patient 24 was deposited to the Japanese Genotype-phenotype Archive under accession number JGAS00000000004.
Reference List
- 1.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bady P, Sciuscio D, Diserens AC, Bloch J, van den Bent MJ, Marosi C, Dietrich PY, Weller M, Mariani L, Heppner FL, Mcdonald DR, Lacombe D, Stupp R, Delorenzi M, Hegi ME. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumert B, Mason WP, Ryan G, Bromberg JE, van den Bent M, Hoang-Xuan K, Brandes AA, Kantor G, Taphoorn MJ, Ben Hassel M, Rees J, Wick W, von DA, Hartmann C, Kros JM, Hegi ME, Dif N, Lacombe D, Gorlia T, Stupp R. The international EORTC/NCIC-CTG/TROG/MRC-CTU low-grade glioma trial: Temozolomide chemotherapy vs. radiotherapy in molecularly characterized (1p loss) LGG. Neuro Oncol. 2013;15 [Google Scholar]
- 4.Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodell WJ, Gaikwad NW, Miller D, Berger MS. Formation of DNA adducts and induction of lacI mutations in Big Blue Rat-2 cells treated with temozolomide: implications for the treatment of low-grade adult and pediatric brain tumors. Cancer Epidemiol Biomarkers Prev. 2003;12:545–551. [PubMed] [Google Scholar]
- 6.Brada M, Viviers L, Abson C, Hines F, Britton J, Ashley S, Sardell S, Traish D, Gonsalves A, Wilkins P, Westbury C. Phase II study of primary temozolomide chemotherapy in patients with WHO grade II gliomas. Ann Oncol. 2003;14:1715–1721. doi: 10.1093/annonc/mdg371. [DOI] [PubMed] [Google Scholar]
- 7.Brandes AA, Franceschi E, Tosoni A, Bartolini S, Bacci A, Agati R, Ghimenton C, Turazzi S, Talacchi A, Skrap M, Marucci G, Volpin L, Morandi L, Pizzolitto S, Gardiman M, Andreoli A, Calbucci F, Ermani M. O(6)-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro Oncol. 2010;12:283–288. doi: 10.1093/neuonc/nop050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB, Batchelor TT, Futreal PA, Stratton MR, Curry WT, Iafrate AJ, Louis DN. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cankovic M, Mikkelsen T, Rosenblum ML, Zarbo RJ. A simplified laboratory validated assay for MGMT promoter hypermethylation analysis of glioma specimens from formalin-fixed paraffin-embedded tissue. Lab Invest. 2007;87:392–397. doi: 10.1038/labinvest.3700520. [DOI] [PubMed] [Google Scholar]
- 10.Chao EC, Velasquez JL, Witherspoon MS, Rozek LS, Peel D, Ng P, Gruber SB, Watson P, Rennert G, Anton-Culver H, Lynch H, Lipkin SM. Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR) Hum Mutat. 2008;29:852–860. doi: 10.1002/humu.20735. [DOI] [PubMed] [Google Scholar]
- 11.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994;14:6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costello JF, Futscher BW, Tano K, Graunke DM, Pieper RO. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269:17228–17237. [PubMed] [Google Scholar]
- 14.Douw L, Klein M, Fagel SS, van den Heuvel J, Taphoorn MJ, Aaronson NK, Postma TJ, Vandertop WP, Mooij JJ, Boerman RH, Beute GN, Sluimer JD, Slotman BJ, Reijneveld JC, Heimans JJ. Cognitive and radiological effects of radiotherapy in patients with low-grade glioma: long-term follow-up. Lancet Neurol. 2009;8:810–818. doi: 10.1016/S1474-4422(09)70204-2. [DOI] [PubMed] [Google Scholar]
- 15.Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, Crooks D, Husband D, Shenoy A, Brodbelt A, Wong H, Liloglou T, Haylock B, Walker C. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101:124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 17.Everhard S, Tost J, El AH, Criniere E, Busato F, Marie Y, Gut IG, Sanson M, Mokhtari K, Laigle-Donadey F, Hoang-Xuan K, Delattre JY, Thillet J. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11:348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsberg J, Thon N, Eigenbrod S, Hentschel B, Sabel MC, Westphal M, Schackert G, Kreth FW, Pietsch T, Loffler M, Weller M, Reifenberger G, Tonn JC. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129:659–670. doi: 10.1002/ijc.26083. [DOI] [PubMed] [Google Scholar]
- 19.Fritz G, Kaina B. Genomic differences between O6-methylguanine-DNA methyltransferase proficient (Mex+) and deficient (Mex-) cell lines: possible role of genetic and epigenetic changes in conversion of Mex+ into Mex- Biochem Biophys Res Commun. 1992;183:1184–1190. doi: 10.1016/s0006-291x(05)80315-8. [DOI] [PubMed] [Google Scholar]
- 20.Giraldo A, Gomez A, Salguero G, Garcia H, Aristizabal F, Gutierrez O, Angel LA, Padron J, Martinez C, Martinez H, Malaver O, Florez L, Barvo R. MLH1 and MSH2 mutations in Colombian families with hereditary nonpolyposis colorectal cancer (Lynch syndrome)--description of four novel mutations. Fam Cancer. 2005;4:285–290. doi: 10.1007/s10689-005-4523-7. [DOI] [PubMed] [Google Scholar]
- 21.Grasbon-Frodl EM, Kreth FW, Ruiter M, Schnell O, Bise K, Felsberg J, Reifenberger G, Tonn JC, Kretzschmar HA. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J Cancer. 2007;121:2458–2464. doi: 10.1002/ijc.23020. [DOI] [PubMed] [Google Scholar]
- 22.Groenendijk FH, Taal W, Dubbink HJ, Haarloo CR, Kouwenhoven MC, van den Bent MJ, Kros JM, Dinjens WN. MGMT promoter hypermethylation is a frequent, early, and consistent event in astrocytoma progression, and not correlated with TP53 mutation. J Neurooncol. 2011;101:405–417. doi: 10.1007/s11060-010-0274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton MG, Roldan G, Magliocco A, McIntyre JB, Parney I, Easaw JC. Determination of the methylation status of MGMT in different regions within glioblastoma multiforme. J Neurooncol. 2011;102:255–260. doi: 10.1007/s11060-010-0307-5. [DOI] [PubMed] [Google Scholar]
- 24.Harris LC, Remack JS, Brent TP. Identification of a 59 bp enhancer located at the first exon/intron boundary of the human O6-methylguanine DNA methyltransferase gene. Nucleic Acids Res. 1994;22:4614–4619. doi: 10.1093/nar/22.22.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de TN, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 26.Hunter C, Smith R, Cahill DP, Stephens P, Stevens C, Teague J, Greenman C, Edkins S, Bignell G, Davies H, O'Meara S, Parker A, Avis T, Barthorpe S, Brackenbury L, Buck G, Butler A, Clements J, Cole J, Dicks E, Forbes S, Gorton M, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Kosmidou V, Laman R, Lugg R, Menzies A, Perry J, Petty R, Raine K, Richardson D, Shepherd R, Small A, Solomon H, Tofts C, Varian J, West S, Widaa S, Yates A, Easton DF, Riggins G, Roy JE, Levine KK, Mueller W, Batchelor TT, Louis DN, Stratton MR, Futreal PA, Wooster R. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–3991. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeltsch A. Bisulfite Sequencing DNA Methylation Analysis; analysis of primary bisulfite sequencing DNA methylation data. 2013 [Google Scholar]
- 28.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 29.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, Nelson SJ, Bollen AW, Gustafson WC, Charron E, Weiss WA, Smirnov IV, Song JS, Olshen AB, Cha S, Zhao Y, Moore RA, Mungall AJ, Jones SJ, Hirst M, Marra MA, Saito N, Aburatani H, Mukasa A, Berger MS, Chang SM, Taylor BS, Costello JF. Mutational Analysis Reveals the Origin and Therapy-Driven Evolution of Recurrent Glioma. Science. 2013 doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koekkoek JA, Dirven L, Heimans JJ, Postma TJ, Vos MJ, Reijneveld JC, Taphoorn MJ. Seizure reduction in a low-grade glioma: more than a beneficial side effect of temozolomide. J Neurol Neurosurg Psychiatry. 2014 doi: 10.1136/jnnp-2014-308136. [DOI] [PubMed] [Google Scholar]
- 31.Komine C, Watanabe T, Katayama Y, Yoshino A, Yokoyama T, Fukushima T. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression free survival in patients with low-grade diffuse astrocytomas. Brain Pathol. 2003;13:176–184. doi: 10.1111/j.1750-3639.2003.tb00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malley DS, Hamoudi RA, Kocialkowski S, Pearson DM, Collins VP, Ichimura K. A distinct region of the MGMT CpG island critical for transcriptional regulation is preferentially methylated in glioblastoma cells and xenografts. Acta Neuropathol. 2011;121:651–661. doi: 10.1007/s00401-011-0803-5. [DOI] [PubMed] [Google Scholar]
- 35.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura M, Watanabe T, Yonekawa Y, Kleihues P, Ohgaki H. Promoter methylation of the DNA repair gene MGMT in astrocytomas is frequently associated with G:C -->A:T mutations of the TP53 tumor suppressor gene. Carcinogenesis. 2001;22:1715–1719. doi: 10.1093/carcin/22.10.1715. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen SA, Stechishin OD, Luchman HA, Lun XQ, Senger DL, Robbins SM, Cairncross G, Weiss S. Novel MSH6 mutations in treatment naive glioblastoma and anaplastic oligodendroglioma influence temozolomide resistance independently of MGMT methylation. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-13-1856. [DOI] [PubMed] [Google Scholar]
- 38.Pace A, Vidiri A, Galie E, Carosi M, Telera S, Cianciulli AM, Canalini P, Giannarelli D, Jandolo B, Carapella CM. Temozolomide chemotherapy for progressive low-grade glioma: clinical benefits and radiological response. Ann Oncol. 2003;14:1722–1726. doi: 10.1093/annonc/mdg502. [DOI] [PubMed] [Google Scholar]
- 39.Parkinson JF, Wheeler HR, Clarkson A, McKenzie CA, Biggs MT, Little NS, Cook RJ, Messina M, Robinson BG, McDonald KL. Variation of O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation in serial samples in glioblastoma. J Neurooncol. 2008;87:71–78. doi: 10.1007/s11060-007-9486-0. [DOI] [PubMed] [Google Scholar]
- 40.Peltomaki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174–1179. doi: 10.1200/JCO.2003.04.060. [DOI] [PubMed] [Google Scholar]
- 41.Pieper RO, Costello JF, Kroes RA, Futscher BW, Marathi U, Erickson LC. Direct correlation between methylation status and expression of the human O-6-methylguanine DNA methyltransferase gene. Cancer Commun. 1991;3:241–253. doi: 10.3727/095535491820873092. [DOI] [PubMed] [Google Scholar]
- 42.Pignatti F, van den Bent M, Curran D, Debruyne C, Sylvester R, Therasse P, Afra D, Cornu P, Bolla M, Vecht C, Karim AB. Prognostic factors for survival in adult patients with cerebral low-grade glioma. J Clin Oncol. 2002;20:2076–2084. doi: 10.1200/JCO.2002.08.121. [DOI] [PubMed] [Google Scholar]
- 43.Quinn JA, Reardon DA, Friedman AH, Rich JN, Sampson JH, Provenzale JM, McLendon RE, Gururangan S, Bigner DD, Herndon JE, Avgeropoulos N, Finlay J, Tourt-Uhlig S, Affronti ML, Evans B, Stafford-Fox V, Zaknoen S, Friedman HS. Phase II trial of temozolomide in patients with progressive low-grade glioma. J Clin Oncol. 2003;21:646–651. doi: 10.1200/JCO.2003.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Raevaara TE, Korhonen MK, Lohi H, Hampel H, Lynch E, Lonnqvist KE, Holinski-Feder E, Sutter C, McKinnon W, Duraisamy S, Gerdes AM, Peltomaki P, Kohonen-Ccorish M, Mangold E, Macrae F, Greenblatt M, de la Chapelle A, Nystrom M. Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology. 2005;129:537–549. doi: 10.1016/j.gastro.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Ramalho-Carvalho J, Pires M, Lisboa S, Graca I, Rocha P, Barros-Silva JD, Savva-Bordalo J, Mauricio J, Resende M, Teixeira MR, Honavar M, Henrique R, Jeronimo C. Altered expression of MGMT in high-grade gliomas results from the combined effect of epigenetic and genetic aberrations. PLoS One. 2013;8:e58206. doi: 10.1371/journal.pone.0058206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reijneveld JC, Bottomley A, Taphoorn MJ, Coens C, Bromberg JE, Mason, Hoang-Xuan K, Brandes AA, Stupp R, Kantor G, Ben Hassel M, Ryan G, Wick W, Theissen B, Lacombe D, Gorlia T, Baumert B. Health related Quality of Life results from temozolomide chemotherapy vs. radiotherapy in molecularly characterized (1p loss) low-grade glioma. A randomized phase III Intergroup study by the EORTC/NCIC-CTG/TROG/MRC-CTU (EORTC 22033-26033) Neuro Oncol. 2013;15 [Google Scholar]
- 47.Rodriguez-Hernandez I, Garcia JL, Santos-Briz A, Hernandez-Lain A, Gonzalez-Valero JM, Gomez-Moreta JA, Toldos-Gonzalez O, Cruz JJ, Martin-Vallejo J, Gonzalez-Sarmiento R. Integrated analysis of mismatch repair system in malignant astrocytomas. PLoS One. 2013;8:e76401. doi: 10.1371/journal.pone.0076401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA--fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruda R, Magliola U, Bertero L, Trevisan E, Bosa C, Mantovani C, Ricardi U, Castiglione A, Monagheddu C, Soffietti R. Seizure control following radiotherapy in patients with diffuse gliomas: a retrospective study. Neuro Oncol. 2013;15:1739–1749. doi: 10.1093/neuonc/not109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheinin I, Sie D, Bengtsson H, van de Wiel MA, Olshen AB, van Thuijl HF, van Essen HF, Eijk PP, Rustenburg F, Meijer GA, Reijneveld JC, Wesseling P, Pinkel D, Albertson DG, Ylstra B. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw EG, Wang M, Coons SW, Brachman DG, Buckner JC, Stelzer KJ, Barger GR, Brown PD, Gilbert MR, Mehta MP. Randomized trial of radiation therapy plus procarbazine, lomustine, and vincristine chemotherapy for supratentorial adult low-grade glioma: initial results of RTOG 9802. J Clin Oncol. 2012;30:3065–3070. doi: 10.1200/JCO.2011.35.8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin YK, Heo SC, Shin JH, Hong SH, Ku JL, Yoo BC, Kim IJ, Park JG. Germline mutations in MLH1, MSH2 and MSH6 in Korean hereditary non-polyposis colorectal cancer families. Hum Mutat. 2004;24:351. doi: 10.1002/humu.9277. [DOI] [PubMed] [Google Scholar]
- 53.Silber JR, Blank A, Bobola MS, Ghatan S, Kolstoe DD, Berger MS. O6-methylguanine-DNA methyltransferase-deficient phenotype in human gliomas: frequency and time to tumor progression after alkylating agent-based chemotherapy. Clin Cancer Res. 1999;5:807–814. [PubMed] [Google Scholar]
- 54.Taal W, Dubbink HJ, Zonnenberg CB, Zonnenberg BA, Postma TJ, Gijtenbeek JM, Boogerd W, Groenendijk FH, Kros JM, Kouwenhoven MC, van MR, van H I, van der Holt B, Bromberg JE, Sillevis Smitt PA, Dinjens WN, van den Bent MJ. First-line temozolomide chemotherapy in progressive low-grade astrocytomas after radiotherapy: molecular characteristics in relation to response. Neuro Oncol. 2011;13:235–241. doi: 10.1093/neuonc/noq177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van de Wiel MA, Kim KI, Vosse SJ, van Wieringen WN, Wilting SM, Ylstra B. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics. 2007;23:892–894. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- 56.van Thuijl HF, Scheinin I, Sie D, Alentorn A, van Essen HF, Cordes M, Fleischeuer R, Gijtenbeek AM, Beute G, van den Brink WA, Meijer GA, Havenith M, Idbaih A, Hoang-Xuan K, Mokhtari K, Verhaak R, van d V, van de Wiel MA, Heimans JJ, Aronica E, Reijneveld JC, Wesseling P, Ylstra B. Spatial and temporal evolution of distal 10q deletion, a prognostically unfavorable event in diffuse low-grade gliomas. Genome Biol. 2014;15:471. doi: 10.1186/s13059-014-0471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 58.Weber RG, Sabel M, Reifenberger J, Sommer C, Oberstrass J, Reifenberger G, Kiessling M, Cremer T. Characterization of genomic alterations associated with glioma progression by comparative genomic hybridization. Oncogene. 1996;13:983–994. [PubMed] [Google Scholar]
- 59.Wick W, Meisner C, Hentschel B, Platten M, Schilling A, Wiestler B, Sabel MC, Koeppen S, Ketter R, Weiler M, Tabatabai G, von DA, Gramatzki D, Westphal M, Schackert G, Loeffler M, Simon M, Reifenberger G, Weller M. Prognostic or predictive value of MGMT promoter methylation in gliomas depends on IDH1 mutation. Neurology. 2013;81:1515–1522. doi: 10.1212/WNL.0b013e3182a95680. [DOI] [PubMed] [Google Scholar]
- 60. www.clinicaltrials.gov. NCT00003375, Observation or Radiation Therapy With or Without Combination Chemotherapy in Treating Patients With Low-Grade Glioma. 2013
- 61. www.clinicaltrialsregister.eu. EORTC 22022-26033, Evaluating primary chemotherapy with temozolomide continuative low dose in patients with low grade gliomas grade II WHO. 2013
- 62.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yip S, Miao J, Cahill DP, Iafrate AJ, Aldape K, Nutt CL, Louis DN. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15:4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.