Abstract
Background
Precise temporal and spatial expression of the clustered Hox genes is essential for patterning the developing embryo. Temporal activation of Hox genes was shown to be cluster-autonomous. However, gene clustering appears dispensable for spatial colinear expression. Generally, a set of Hox genes expressed in a group of cells instructs these cells about their fate such that the differential expression of Hox genes results in morphological diversity. The spatial colinearity is considered to rely both on local and long-range cis regulation.
Results
Here, we report on the global deregulation of HoxA and HoxD expression patterns upon inactivation of a subset of HOXA and HOXD proteins.
Conclusions
Our data suggest the existence of a “self-regulation” mechanism, a process by which HOX proteins establish and/or maintain the spatial domains of the Hox gene family and we propose that the functionally dominant HOX proteins could contribute to generating the spatial parameters of Hox expression in a given tissue, i.e., HOX controlling the establishment of the ultimate HOX code.
Keywords: Hox genes, transcriptional regulation, limb development
INTRODUCTION
During embryogenesis, developmental events are orchestrated by changes in gene expression. Appropriate spatial and temporal gene expressions execute pattern formation, a process in which cells become sequentially specified and differentiate to form a morphological structure. A gene or subset of genes’ transcriptional outcome, in a cell or group of cells, is regulated at multiple levels, including interactions between promoter, enhancer, transcription factors, epigenetic modifiers and noncoding RNAs. How transcriptional networks are regulated, resulting in complex patterns of gene expression, remain an open question in biology.
The discovery of the Hox cluster in Drosophila revealed an intriguing correspondence between the physical order of the genes within the Hox cluster and their expression domain along the embryonic anterior to posterior (A–P) axis, a phenomenon known as the spatial colinearity (Lewis, 1978). Subsequent studies of orthologous Hox complexes uncovered the conservation of the spatial colinearity in vertebrates and in addition showed that Hox genes are sequentially activated in time from the 3′ to the 5′ end of the cluster (referred to as temporal colinearity) (Gaunt et al., 1986, 1989; Dolle et al., 1989; Duboule and Dolle, 1989; Graham et al., 1989; Izpisua-Belmonte et al., 1991). Spatial and temporal control of Hox gene expression is essential for patterning the vertebrate body plan; yet, the mechanisms underlying spatial and temporal “colinearity” remain largely elusive. After initial activation, spatial expression domains of each Hox gene are progressively refined and further maintained during embryonic development through mechanisms that most likely vary between tissues. The resulting spatial expression domains or the differential qualitative and quantitative combination of different Hox genes products along the axes (referred to as the “HOX Code”) is essential for patterning the developing embryo (Kessel and Gruss, 1991). Therefore, elucidating the mechanisms by which Hox spatial domains are established and maintained is crucial for our understanding of embryonic development and disease.
Genetic analyses in mice suggest that cis-regulatory elements could be implicated in the sequential activation of Hox genes from 3′ to 5′ (Kmita et al., 2002; Spitz et al., 2003; Deschamps, 2007). Sequential posttranslational modifications of histones, i.e., from transcriptionally silent-specific to active-specific forms, also correlate with sequential gene activation (Soshnikova and Duboule, 2009). Furthermore, it was proposed that changes in higher order chromatin organization, e.g., chromatin de-condensation (Noordermeer and Duboule, 2013) and looping out of chromosome territories, contribute to the sequential activation of Hox genes (Chambeyron and Bickmore, 2004; Chambeyron et al., 2005; Morey et al., 2007). Although it remains unclear whether these latter changes are a cause or a consequence of the sequential activation of Hox genes. While sequential activation determines the spatial coordinates for Hox gene expression in some animal phyla, clustering per se appears dispensable for spatial colinearity (Deschamps, 2007; Duboule, 2007). It has thus been proposed that spatial colinearity is an ancestral property whereas clustering and temporal colinearity were imposed during evolution (Duboule, 2007).
During limb development, genes from HoxA and HoxD clusters are activated in a sequential manner following their order within the cluster, leading to expression domains that are colinear both in space and time (Kmita and Duboule, 2003; Zakany and Duboule, 2007; Montavon and Duboule, 2013). Expression of HoxD genes occurs in two independent phases (referred to as phase one and phase two) that rely on distinct cis-regulatory elements located on both sides of the cluster (Nelson et al., 1996; Tarchini and Duboule, 2006; Montavon and Duboule, 2013). The 3′ regulatory region (early limb control region; ELCR) controls the sequential timing of gene activation (phase one expression) (Zakany et al., 2004). Concomitantly, a 5′ regulatory region (POST) exerts a repressive effect to spatially restrict 5′Hoxd expression to the posterior mesenchyme of early limb buds (Tarchini and Duboule, 2006). Subsequently, these early expression domains evolve to later be fixed as the presumptive zeugopod domain (Tarchini and Duboule, 2006). The phase two expressions of HoxD genes occurs exclusively in the presumptive digit-forming region and is mechanistically unlinked to the first expression phase (Spitz et al., 2003; Tarchini and Duboule, 2006; Gonzalez et al., 2007; Montavon et al., 2011). Distinct remote regulatory elements, located in the gene desert on the 5′ side of the cluster, drive the phase two transcription (Montavon et al., 2011; Tschopp and Duboule, 2011). This occurs in a reverse colinear manner as the most 5′transcription unit, i.e., Hoxd13, is expressed strongly in the entire presumptive digit territory while Hoxd12 to Hoxd9 are transcribed with progressively lower efficiency and are excluded from the digit-1 region (Kmita et al., 2002; Montavon et al., 2008). In both phases, the position of transcription unit relative to the regulatory regions determines the spatial and temporal parameters of HoxD expression. Changes in the genomic position of a transcription unit relative to the 3′ regulatory region (e.g., by targeted deletion in the cluster) alter its time of activation, while spatial expression is affected depending on its relative position to the 5′ regulatory regions. It is believed that similar mechanisms apply for the HoxA cluster.
In this study, through the analysis of different HoxA and HoxD mutants, we provide evidence that HOX proteins themselves are important to set up the spatial parameters of HoxA and HoxD expression in distal limb buds. As previously reported, we observed that deletion of 5′Hoxd genes affects only HoxD expression. In contrast, loss of HOXA13 protein has a more general impact upon the expression of both HoxA and HoxD genes. Furthermore, we show that presence of the HOX paralogous group 13 proteins is a prerequisite for the separation of “zeugopod” and “autopod” expression domains of HoxA and HoxD genes.
RESULTS
Expression of the Remaining Hox Genes in Limb Buds of HoxDdel(11–13)/del(11–13) Embryos
The HoxDdel(11–13) allele used in this study carries the deletion in cis of the Hoxd13 and Hoxd12 loci, as well as the insertion of a lacZ reporter transgene within the first exon of the Hoxd11 gene (i.e., combined loss of function of Hoxd13, 12, and 11) (Zakany and Duboule, 1996). This deletion modifies the position of the remaining transcription units relative to the 5′ enhancers, such that Hoxd11-lacZ is relocated at the former position of Hoxd13. Based on our current understanding of HoxD genes regulation, deletion of the 5′Hoxd genes should not alter the time of activation of the remaining Hoxd genes, as their position relative to the 3′ regulatory region (ELCR) is unchanged (Zakany et al., 2004; Tarchini and Duboule, 2006). In contrast, this deletion brings the remaining HoxD genes closer to the 5′ regulatory landscape, which is expected to posteriorly restrict their phase one expression in the early limb bud and subsequently modify their expression in the presumptive zeugopod domain (e.g., Hoxd10 should be expressed like Hoxd12). In addition, it has been shown that 5′ deletion within the HoxD cluster affects the phase two expression of only the first transcription unit nearest to the deletion site (Kmita et al., 2002), i.e., Hoxd11-LacZ should be expressed like Hoxd13 without affecting the remaining Hoxd genes.
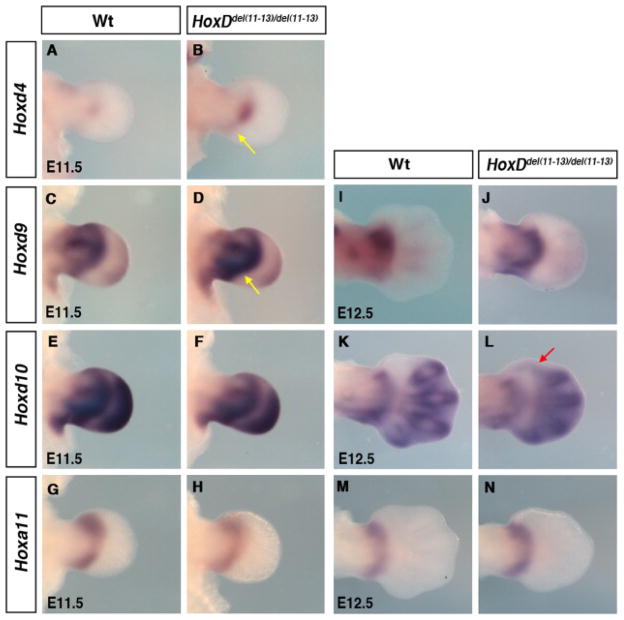
Therefore, we first checked the effect of the deletion on Hoxd4, Hoxd9 and Hoxd10 expression at embryonic day (E) 11.5, when the presumptive zeugopod domain (phase one) and the presumptive autopod domain (phase two) can be simultaneously observed. At E11.5, Hoxd4 was expressed in a small central domain in the wild-type limb bud (Fig. 1A), whereas its domain slightly extended into the posterior limb mesenchyme in HoxDdel(11–13) homozygote embryos (yellow arrow in Fig. 1B). At this stage, while the presumptive zeugopod domain of Hoxd9 shows an anterior bias in wild-type limb buds (Fig. 1C), a posterior bias is observed in HoxDdel(11–13) homozygote limb buds (yellow arrow in Fig. 1D). No difference is observed for Hoxd10 expression in the presumptive zeugopod, which is already posteriorly biased in wild-type limb buds (Fig. 1E,F). The pattern of Hoxd9 and Hoxd10 observed in mutant limb buds at E11.5 is maintained at E12.5 (Fig. 1J,L). Phase two expression of Hoxd9 and Hoxd10 in the presumptive digit-forming region is mostly unaffected by the deletion both at E11.5 and E12.5 (Fig. 1C–F,I–L). Nonetheless, there is an unexpected ectopic anterior expansion of Hoxd10 expression into the presumptive digit-1 territory (red arrow in Fig. 1L), even though Hoxd10 is not adjacent to the deletion breakpoint. No alteration of HoxA genes expression has been reported in the absence of HoxD genes. Accordingly, no modification of Hoxa13 expression is observed in HoxDdel(11–13) homozygous limb buds (Sheth et al., 2007). Similarly, Hoxa11 expression is unchanged in HoxDdel(11–13) homozygous limb buds and remains restricted to the zeugopod both at E11.5 (Fig. 1G,H) and E12.5 (Fig. 1M,N).
Fig. 1.
Effect of HoxDdel(11–13) deletion on Hox genes expression. A–M: Limb buds hybridized with Hoxd4 (A,B), Hoxd9 (C,D), Hoxd10 (E,F), and Hoxa11 (G,H) at embryonic day (E) 11.5 and Hoxd9(I,J), Hoxd10(K,L), and Hoxa11 (M,N) at E12.5. B,D,J: Yellow arrow points at posteriorly biased expression of Hoxd4 (B) and Hoxd9 (D,J). L: Red arrow point at ectopic second phase expression of Hoxd10 in digit-1 region. Note that the slight difference in the morphology of HoxDdel(11–13)/del(11–13) presumptive autopod at E12.5 prefigures the eventual skeletal phenotype of HoxDdel(11–13) homozygous mice, i.e., shortening of digits and up to six digits per limb.
Loss of HOXA13 Affects HoxA and HoxD Expression in the Presumptive Zeugopod Domain
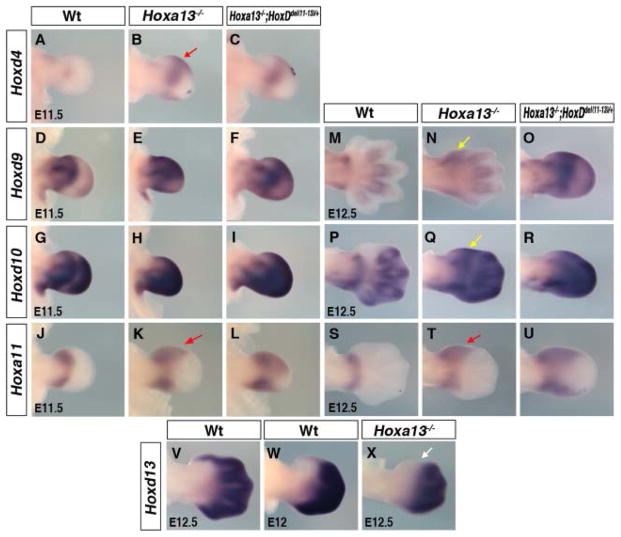
A few examples of cross-regulatory interactions among Hox genes have been reported but this is not considered as a general mechanism of Hox gene regulation (Arcioni et al., 1992; Popperl et al., 1995; Studer et al., 1998; Manzanares et al., 2001). It has been suggested that HOXA13 may participate in transcriptional repression of Hoxa11, and shown that Hoxa11 expression marginally extends into the presumptive digit-1 region of Hoxa13 mutant limb buds (Yokouchi et al., 1995; Post and Innis, 1999). Surprisingly, in Hoxa13 mutant limb buds, in which the Homeobox containing region of Hoxa13 is disrupted (Fromental-Ramain et al., 1996), we found that Hoxd4 expression is upregulated and distally expanded in the anterior and posterior mesenchyme (Fig. 2A,B, red arrow in Fig. 2B). In addition, Hoxd9 (Fig. 2D,E and 2M,N), Hoxd10 (Fig. 2G,H and 2P,Q), and Hoxd11 (not shown) transcripts are found in the presumptive digit-1 region (yellow arrow in Fig. 2N and 2Q), from which they are excluded in wild-type limb buds (Fig. 2M,P). Similar ectopic expression of these Hoxd genes is observed in Hoxa13−/−;HoxDdel(11–13)/+ limb buds (Fig. 2C,F,I,O,R and not shown). The ectopic expression of Hoxd4, Hoxd9, Hoxd10, and Hoxd11 in mutant limb buds is accompanied by an expansion of Hoxa11 expression in that region (Fig. 2J–L and 2S–U, red arrow in 2K and 2T).
Fig. 2.
Deregulation of Hox genes expression in Hoxa13−/− and Hoxa13−/−;HoxDdel(11–13/+ mutant limb buds. A–X: Limb buds hybridized with Hoxd4 (A–C), Hoxd9 (D–F), Hoxd10 (G–I), and Hoxa11 (J–L) at embryonic day (E) 11.5, and Hoxd9 (M–O), Hoxd10 (P–R), Hoxa11 (S–U), Hoxd13 (V–X) at E12.5. B,C,E,F,H,I: Note deregulation of Hoxd4 (B,C), Hoxd9 (E,F) and Hoxd10 (H,I) expression. N,Q: The yellow arrow points at ectopic first phase expression of Hoxd9 (N) and Hoxd10 (Q) in presumptive digit-1 region. B,K,T: The red arrow points at the anterior distal extension of Hoxd4 (B) and Hoxa11 (K,T) expression. V–X: Note that Hoxd13, which is normally expressed in presumptive digit-1 (V,W), is excluded from this region in Hoxa13 mutants (white arrow in X). The difference in the shape of the presumptive digit domains of Hoxa13−/− and Hoxa13−/−; HoxDdel11–13/+ embryos at E12.5 prefigures the eventual oligodactyl phenotype.
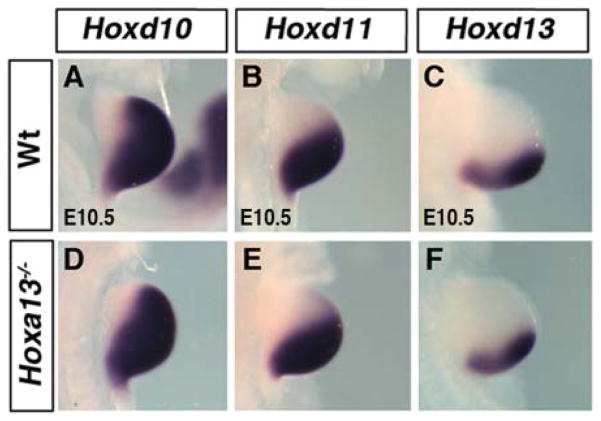
The presumptive zeugopod domain of HoxD genes at E11.5 and E12.5 has been suggested to be the continuation of phase one regulation (Tarchini and Duboule, 2006). In this view, the ectopic expressions described above, which look like an extension of the presumptive zeugopod domain, could reflect a modification of the phase one expression in early buds resulting from Hoxa13 inactivation. However, Hoxa13 expression starts at E10.5 in a small distal-posterior domain, mostly nonoverlapping with the phase one expression. Moreover, Hoxd10, Hoxd11, and Hoxd13 expression does not show any alteration of phase one expression in early limb buds of Hoxa13−/− embryos (Fig. 3). Therefore, the effect of Hoxa13 loss of function on HoxD expression is most likely unrelated to the phase one regulation in early limb bud but rather reflects the role of HOXA13 protein in preventing the phase one expression in the presumptive autopod (mesopod and acropod) domain. Accordingly, while the phase one and phase two expression domains of Hoxd9, Hoxd10, and Hoxd11 are normally separated by a stripe of cells devoid of either transcripts, this gap is reduced in Hoxa13−/− and Hoxa13−/−; HoxDdel(11–13)/+ limb buds (Fig. 2D–I, 2M–R and not shown). This reduction in the gap between phase one and phase two expression domains may be secondary to growth defect in the distal region. However, the broadening of Hoxa11 expression pattern in Hoxa13−/− and Hoxa13−/−; HoxDdel(11–13)/+ limb buds (Fig. 2S–U) suggest a deregulation of HoxD expression in the autopod territory rather than reduced growth in the mesopod region. This is further supported by the virtually normal expression of Hoxa13-exon1 in mutant limb buds (see below). Altogether, these results suggest a mechanism whereby HOXA13 protein suppresses the expression of Hoxa11 and phase one regulation of HoxD genes in the presumptive mesopod-acropod territory.
Fig. 3.
Expression of 5′Hoxd genes in Hoxa13−/− limb bud. Embryonic day (E) 10.5 forelimb buds hybridized with Hoxd10 (A,D), Hoxd11 (B,E), and Hoxd13 (C,F). Note that the phase one expression of 5′Hoxd genes is unaltered in absence of Hoxa13. Genotypes are indicated on the left.
Paralogous HOX proteins are characterized by some redundancy in their function (Wellik and Capecchi, 2003). In addition to Hoxa13, Hoxd13 is the only HoxD gene expressed in the presumptive digit-1 region. Surprisingly, in Hoxa13−/− limb buds, Hoxd13 expression is excluded from digit-1 region (white arrow in Fig. 2X, compared with 2V,W) and its down-regulation correlates with the up-regulation of the other HoxD genes and Hoxa11 (compare Fig. 2N,Q,T with 2X). This raises the possibility that the absence of digit-1 in the Hoxa13 mutant may actually be due to the loss of HOX paralogous group 13 function in this region. In addition, the down-regulation of Hoxd13 in the presumptive digit-1 raises the question of whether Hoxd13 together with Hoxa13 share a similar function in controlling the expression of Hoxa11 and the other HoxD genes.
Cells Destined to Form Autopod are Present in the Absence of 5′ Hox Genes
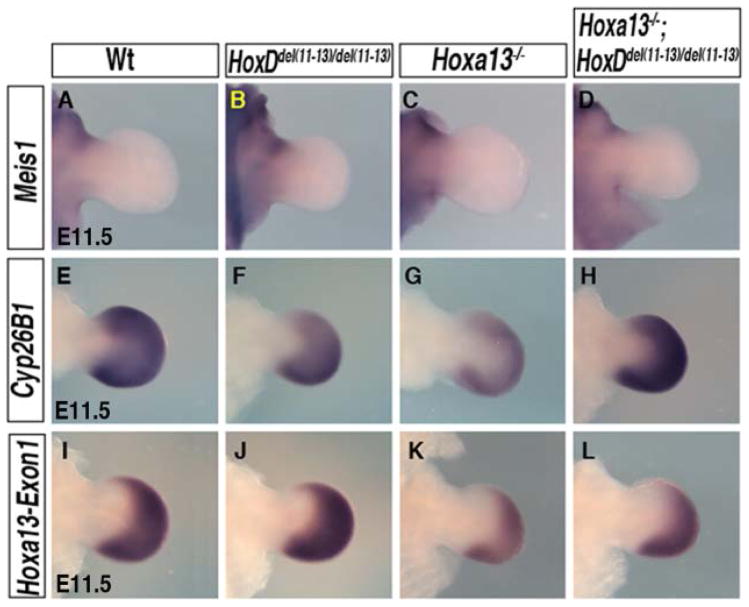
To further assess the functional relevance of 5′HOX products in regulating expression of HoxA and HoxD genes, we decided to analyze the double Hox-a13−/−;HoxDdel(11–13)/del(11–13) mutant. These mutant embryos show complete digit agenesis similar to the Hoxa13−/−;Hoxd13−/− mutant (Fromental-Ramain et al., 1996; Sheth et al., 2012), with no reported defects in the zeugopod and stylopod segments (Zakany et al., 1997). It is frequently assumed that digit loss upon 5′Hox inactivation could be due to the lack of autopod progenitors. Therefore we first tested whether Hoxa13−/−; HoxDdel(11–13)/del(11–13) limb bud cells are properly specified along the proximal–distal (P–D) axis. A proximal signal (presumably retinoic acid) induces Meis1 expression (Cooper et al., 2011; Rosello-Diez et al., 2011), while a distal signal, fibroblast growth factors from the apical ectodermal ridge, regulates expression of the retinoic acid-degrading enzyme Cyp26b1 in the distal mesenchyme (Probst et al., 2011). We thus used Meis1 and Cyp26b1 as markers of proximal and distal cell identities, respectively. In both wild-type and mutant early buds, Meis1 expression gets restricted to the proximal region by E11.5 (Fig. 4A–D), confirming normal specification of the proximal cells. Similarly, Cyp26b1 is expressed with a wild-type pattern in mutant buds (Fig. 4E–H), indicating the normal specification of the distal cells.
Fig. 4.
Proximal and distal limb cells are properly specified in mutant limb buds. A–H: Embryonic day (E) 11.5 forelimb buds show proper specification of proximal limb cells, marked by Meis1 expression (A–D), and of distal cells, marked by Cyp26b1 expression (E–H). I–L: Hoxa13-Exon1 expression marks the cells destined to form autopod.
Hoxa13 is so far the best available marker for specified autopod cells (Tabin and Wolpert, 2007). Its expression starts in the posterior-distal mesenchyme of wild-type forelimb buds around E10.5 and progressively covers the entire digital plate as it forms. The homeobox region of Hoxa13 is disrupted in the Hoxa13−/− mutant but Hoxa13-exon1 remains transcribed (Fromental-Ramain et al., 1996). Therefore, we used Hoxa13-exon1 specific riboprobe to check for the presence of autopod progenitors. In the wild-type limb bud, Hoxa13-exon1 expression is observed in the presumptive autopod and recapitulates the reported Hoxa13 expression pattern. No obvious changes in Hoxa13-exon1 expression is observed in mutant limb buds (Fig. 4I–L), thus indicating that cells destined to form the autopod are present in these mutants. Consequently, Hoxa13−/−; HoxDdel(11–13)/Del(11–13) limb buds provide an appropriate context to study the HOX-dependent regulation of Hox genes in developing limbs.
Function of HOX Paralogous Group 13 is Required for Proper Separation of the Zeugopod and Autopod Expression Domains of HoxA and HoxD Genes
A striking spatial redistribution of HoxA and HoxD transcripts is observed in Hoxa13−/−;HoxDdel(11–13)/Del(11–13) limb buds suggesting a synergistic effect of the combined mutations (Fig. 5). Notably, Hoxd4 is ectopically expressed in the distal mesenchyme (Fig. 5B) and the separation between the phase one and two expression of Hoxd9 and Hoxd10 is completely lost (Fig. 5D,F compared with 5C,E). In addition, Hoxa11 expression, which marks the boundary between the prospective zeugopod and autopod in wild-type limbs, expands distally to cover the entire presumptive autopod region of the mutant limb bud (Fig. 5H compared with 5G). Altogether, these modifications indicate that the presence of 5′HOX products is required first to suppress Hoxa11 and the phase one expression of HoxD genes in the presumptive autopod domain and second for the proper separation of the presumptive zeugopod and autopod expression domains.
Fig. 5.
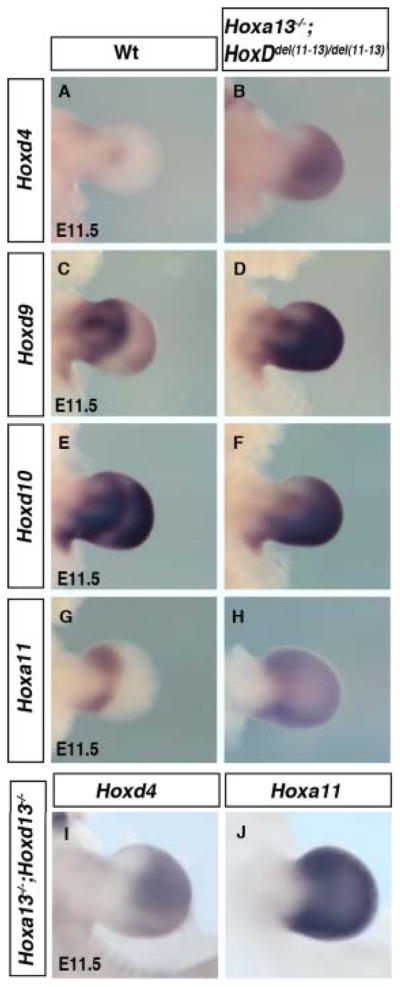
Spatial redistribution of remaining Hox genes in Hoxa13−/−;HoxDdel(11–13/del(11–13) and Hoxa13−/−;Hoxd13−/−. A–J: Embryonic day (E) 11.5 forelimb buds hybridized with Hoxd4 (A,B,I), Hoxd9 (C,D), Hoxd10 (E,F), and Hoxa11 (G,H,I). B,H–J: Note similar distal expansion of Hoxd4 (B,I) and Hoxa11 (H,J) in both genotypes.
The most 5′Hox genes are functionally dominant when co-expressed with other Hox genes, a phenomenon described as posterior prevalence (Duboule and Morata, 1994). Therefore, to test whether the observed effects are primarily due to loss of HOX paralogous group 13 proteins, we analyzed mutant limb buds in which HOXA13 and HOXD13 are inactivated. This also permits the evaluation of the possible contribution of HOXD11 and HOXD12. Our results show that Hoxd4 and Hoxa11 expression is extended distally and covers the entire distal limb bud in Hoxa13−/−;Hoxd13−/− mutants (Fig. 5I,J), recapitulating the situation in Hoxa13−/−;HoxDdel(11–13)/del(11–13) buds (Fig. 5B,H). This confirms that the modified HoxD and Hoxa11 expression in Hoxa13−/−; HoxDdel(11–13)/del(11–13) buds is actually due to the lack of HOXA13 and HOXD13 proteins. These results, for the first time, show that the function of HOX paralogous group 13 is required for the correct spatial expression of the genes from both the HoxA and HoxD clusters.
DISCUSSION
During limb development, proximal cells, marked by Meis1 expression, are fated to form the stylopod (arm), while more distal mesenchyme eventually gives rise to two distinct anatomical segments: the zeugopod (forearm) and the autopod that includes the mesopod (wrist) and the acropod (digits). The distal mesenchyme is molecularly subdivided by the expression of HoxA and HoxD genes, with Hoxa11 expressed in the presumptive zeugopod region while Hoxa13 is expressed in the presumptive mesopod and acropod region. The biphasic regulation of HoxD genes triggers expression in the presumptive zeugopod region (phase one) and acropod region (phase two). A fundamental question is how this molecular subdivision, which is specific to tetrapod limbs, is generated and maintained.
Alterations in HoxA and HoxD expression have been observed in limb buds of different mutants. Curiously, most of these alterations are along the A–P axis, while alterations along the P–D axis have been reported only for HoxD genes upon cis-deletions (Kmita et al., 2002; Tarchini and Duboule, 2006). The changes in HoxA and HoxD expression reported in this work is, to our knowledge, the first evidence for the most 5′ HOX products controlling the expression of both HoxA and HoxD genes along the P–D axis.
Function of HOX Paralogous Group 13 Proteins is Required for the Segregation of Hox Domains Along the P–D Axis
Our results reveal that the deletion of the three most 5′Hoxd genes, in cis, besides affecting the expression of the HoxD gene adjacent to the deletion breakpoint (Zakany and Duboule, 1996), also has some previously unappreciated effects. In the HoxDdel(11–13) allele, Hoxd4 and Hoxd9 are separated from the deletion breakpoint by four and two transcription units, respectively, and their expression is thus expected to remain as in wild-type buds. Yet, we found that their expression in the presumptive zeugopod domain is posteriorly biased in the mutant context. This may be the consequence of their modified position with respect to the 5′ repressive region (POST), which in turn would suggest that POST exert its effect over longer distance than previously reported (Tarchini and Duboule, 2006). However, such change in expression is not observed for Hoxd10 even though it is located closer to the deletion breakpoint than Hoxd9 and Hoxd4. Of interest, a recent report shows that deletion of the 5′ regulatory landscape only affects the phase two expression of 5′Hoxd genes (Hoxd10–12), while the presumptive zeugopod domains remain unaffected (Montavon et al., 2011). Together these results suggest that the establishment of presumptive zeugopod expression domains may be differently regulated for Hoxd1–9 than for Hoxd10–12. Nonetheless, in this mutant background, the biphasic expression of HoxD genes and the segregation of phase one and phase two of HoxD are maintained and the expression of HoxA genes is unaffected.
In contrast to HoxDdel(11–13)/del(11–13), inactivation of Hoxa13 results in two main changes: (i) Hoxa11 and HoxD genes are ectopically expressed distally, mainly in the presumptive digit-1 territory and (ii) separation between phase one and phase two expression domains is significantly reduced. These results suggest that the mechanism that suppresses phase one regulation of HoxD genes and Hoxa11 expression in the presumptive mesopod and digit-1 region is impaired upon Hoxa13 inactivation. This indicates that HOXA13 may play a critical role in segregating Hox expression in the presumptive zeugopod and auto-pod domains. Furthermore, our results show that HOXA13 and HOXD13 act in a synergistic manner as when both proteins are inactivated, Hoxa11 and Hoxd4 transcription occurs in the entire distal limb bud and segregation of the zeugopod and autopod domains of HoxD genes is lost. Together our data provide compelling evidence that HOX paralogous group 13 proteins are crucial for the proper segregation of Hox domains along the P–D axis.
Even though HOX13 proteins act in a synergistic manner (Fromental-Ramain et al., 1996), our results indicate a prominent role for HOXA13. Most of the global change in regulation is observed only when Hoxa13 is inactivated while the effect of the 5′HoxD deletion is only detectable for the remaining HoxD genes. The predominant effect of Hoxa13 could be due to its expression pattern, which covers the entire autopod and mesopod domain more rapidly than Hoxd13, rather than differences in HOXA13 and HOXD13 functional properties per se.
Possible Mechanisms by Which HOX Proteins Contribute to Cluster-wide Regulation
The absence of HOX paralogous group 13 proteins results in ectopic expression of Hoxa11 and Hoxd4 distally and loss of segregation of the zeugopod and autopod expression domains of HoxD genes. As far as HoxD genes are concerned, several possibilities could account for their deregulation: (i) the phase one regulation is ectopically active in the presumptive autopod cells; (ii) phase two regulation loses its restriction to the most 5′ genes, thereby triggering ectopic expression of more 3′genes in the presumptive autopod; and (iii) a combination of the two latter scenarios. Based on the progressive distalization of Hoxd4 expression in the mutant contexts, we favor the ectopic activity of phase one in autopod cells as being responsible for the HoxD deregulation in absence of HOX paralogous group 13 proteins. However, a detailed study of chromatin organization of the HoxD locus in the mutant contexts is needed to unambiguously resolve which phase of regulation is affected. Nonetheless, our results reveal that the function of HOX paralogous group 13 proteins, in particularly HOXA13, is crucial to establish the “dichotomy” of HoxD expression (zeugopod and autopod domains), thus generating an intermediate HoxD-less domain considered as the presumptive wrist domain (Woltering and Duboule, 2010)
A very recent study, identified a regulatory domain located 3′ to the HoxD cluster (T-DOM) controlling the phase one expression of HoxD genes in the early limb bud and presumptive zeugopod (Andrey et al., 2013), while the second phase of expression is controlled by the regulatory domain located 5′ to the HoxD cluster (C-DOM) (Montavon et al., 2011; Andrey et al., 2013). The switch from phase one to phase two regulation in distal cells is accompanied by posttranslational histone modifications, switching the chromatin state from active to inactive at the T-DOM and from inactive to active at the C-DOM. Of interest, the silencing of the phase one regulation in distal cells is independent of phase two activation and it was proposed that the gap between HoxD zeugopod and autopod expression domains corresponds to cells in which phase one is switch off but phase two is not activated (Andrey et al., 2013). Based on our results revealing the loss of this gap in absence of HOX paralogous group13 proteins, we propose that these proteins, may act as a switch to turn-off the phase one regulation. The evidence that HOX proteins can interact with histone/chromatin modifying complexes (Shen et al., 2001; Lu et al., 2003; Luke et al., 2006), raises the possibility that HOX paralogous group 13 proteins may influence the chromatin state at the HoxD cluster and/or its regulatory landscapes. Alternatively, HOX paralogous group 13 proteins could induce minor changes in the chromatin conformation at the HoxD locus disrupting its interaction with phase one regulatory sequences while favoring the interaction underlying phase two regulation in the presumptive autopod domain.
Random Incident or a Coordinate Strategy?
Over the years, Hox genes have been studied either as individual genes or as a cluster. Only in few cases, such as during hindbrain patterning, “local” auto- or cross-regulatory interactions among Hox genes have been reported (Arcioni et al., 1992; Popperl et al., 1995; Studer et al., 1998; Manzanares et al., 2001). Here, we show that the HOX proteins themselves are likely involved in generating “global” spatial parameters of Hox expression in developing limbs. We propose that the establishment of Hox expression patterns involves a “self-regulatory” mechanism whereby functionally dominant HOX proteins determine the spatial parameters of the other Hox genes’ expression in a given tissue. In this view, in animals, in which Hox genes are either clustered or non-clustered, the functionally dominant HOX protein ultimately establishes and/or maintains the spatial expression specificities or HOX code, eventually defining cell fate.
Simple Step and Big Leap Forward
The appearance of paired appendages was a major step in tetrapod evolution and land colonization (Coates, 1994; Clack, 2005). Fossil data suggest that limbs evolved from fins, but how this morphological transformation occurred is not yet resolved (Cohn et al., 2002; Schneider and Shubin, 2013). Curiously, HoxA and HoxD cluster genes are expressed in overlapping domains in fins while the hallmark of tetrapod limbs is the segregation of Hox expression domains in the distal limb bud, at least in the species studied so far (Metscher et al., 2005; Woltering and Duboule, 2010). Segregation of Hox expression domains has been suggested as an important change in the evolution of well-articulated, functional tetrapod limbs (Metscher et al., 2005; Woltering and Duboule, 2010).
The acquisition of new cis-regulatory elements modulating Hox expression or increased distal cell proliferation by prolonged AER function have been suggested as possible mechanisms involved in this evolutionary transformation (Sordino et al., 1995; Freitas et al., 2012; Schneider and Shubin, 2013). Recent studies in mice show that HoxA and HoxD genes are important for limb bud growth in addition to patterning (Kmita et al., 2005; Sheth et al., 2013). HOXD13 overexpression in zebrafish fins results in distal overgrowth, segregation of Hox domains and expression of markers specific to autopod (Freitas et al., 2012). Here we show that HOX13 proteins are important for the segregation of the other Hox domains along the P–D axis. Upon inactivation of HOX13, the other HoxA and HoxD genes are expressed in overlapping domains resembling Hox expression patterns in fish-fins. Therefore, we speculate that, in the course of evolution, acquisition of new cis-regulatory elements and/or modulation of AER signaling contributed to the distal specific expression of Hoxa13. In turn, distal expression of Hoxa13 ensured the expansion of autopod progenitors and generated the conditions required for segregation of the autopod and zeugopod domains of HoxA and HoxD genes, allowing for the development of the wrist and digits and thus giving rise to well-articulated, functional tetrapod limbs.
EXPERIMENTAL PROCEDURES
Mice
The Hoxa13 mutant allele is a generated by insertion of neomycine cassette in Hoxa13 coding region (Fromental-Ramain et al., 1996). The HoxDdel(11–13) allele is the deletion of Hoxd13,Hoxd12 loci plus the insertion of lacZ reporter transgene in Hoxd11 therefore represents Hoxd11–13 loss of function (Zakany and Duboule, 1996). In Hoxd13 mutant allele, a lacZ reporter transgene is inserted in the first exon of Hoxd13 (Kmita et al., 2000). The Hoxa13, HoxDdel(11–13)_ and Hoxd13 mutant lines were maintained in a mixed background.
Noon of the day the vaginal plug was observed was considered as E0.5. The embryos were obtained by caesarean and genotyping was performed by PCR as described.
In Situ Hybridization
Digoxigenin-labeled antisense riboprobe were prepared, and whole-mount in situ hybridization was performed according to standard procedure (Sheth et al., 2007). The probes used were Hoxd4, Hoxd9, Hoxd10, Hoxd11, Hoxd13, and HoxA11 (kindly provided by D. Duboule) and Hoxa13-exon1 (kindly provided by S. Stadler) and Meis1 and Cyp26b1 (kindly provided by M. Torres).
Acknowledgments
Grant sponsor: the Spanish Government; Grant number: BFU2011-24972; Grant sponsor: the Canadian Institutes for Health Research; Grant number: MOP-82880; Grant number: 126110.
We thank Denis Duboule and Pierre Chambon for providing the mutant mice. This work was supported by the Spanish Government to M.R. and by the Canadian Institutes for Health Research as well as the Canada Research Chair program to M.K. R.S was supported by a Formación Profesorado Universitario fellowship from the Spanish Ministry of Science and Innovation and currently supported by the Angelo Pizzagalli postdoctoral fellowship.
References
- Andrey G, Montavon T, Mascrez B, Gonzalez F, Noordermeer D, Leleu M, Trono D, Spitz F, Duboule D. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340:1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- Arcioni L, Simeone A, Guazzi S, Zappavigna V, Boncinelli E, Mavilio F. The upstream region of the human homeobox gene HOX3D is a target for regulation by retinoic acid and HOX homeoproteins. EMBO J. 1992;11:265–277. doi: 10.1002/j.1460-2075.1992.tb05049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA. Nuclear reorganisation of the Hoxb complex during mouse embryonic development. Development. 2005;132:2215–2223. doi: 10.1242/dev.01813. [DOI] [PubMed] [Google Scholar]
- Clack JA. Getting a leg up on land. Sci Am. 2005;293:100–107. doi: 10.1038/scientificamerican1205-100. [DOI] [PubMed] [Google Scholar]
- Coates MI. The origin of vertebrate limbs. Dev Suppl. 1994:169–180. [PubMed] [Google Scholar]
- Cohn MJ, Lovejoy CO, Wolpert L, Coates MI. Branching, segmentation and the metapterygial axis: pattern versus process in the vertebrate limb. Bioessays. 2002;24:460–465. doi: 10.1002/bies.10088. [DOI] [PubMed] [Google Scholar]
- Cooper KL, Hu JK, ten Berge D, Fernandez-Teran M, Ros MA, Tabin CJ. Initiation of proximal-distal patterning in the vertebrate limb by signals and growth. Science. 2011;332:1083–1086. doi: 10.1126/science.1199499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschamps J. Ancestral and recently recruited global control of the Hox genes in development. Curr Opin Genet Dev. 2007;17:422–427. doi: 10.1016/j.gde.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Dolle P, Izpisua-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–364. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Freitas R, Gomez-Marin C, Wilson JM, Casares F, Gomez-Skarmeta JL. Hoxd13 contribution to the evolution of vertebrate appendages. Dev Cell. 2012;23:1219–1229. doi: 10.1016/j.devcel.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996;122:2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ, Miller JR, Powell DJ, Duboule D. Homoeobox gene expression in mouse embryos varies with position by the primitive streak stage. Nature. 1986;324:662–664. doi: 10.1038/324662a0. [DOI] [PubMed] [Google Scholar]
- Gaunt SJ, Krumlauf R, Duboule D. Mouse homeo-genes within a subfamily, Hox-1.4, ′2.6 and ′5.1, display similar anteroposterior domains of expression in the embryo, but show stage- and tissue-dependent differences in their regulation. Development. 1989;107:131–141. doi: 10.1242/dev.107.1.131. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Duboule D, Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev Biol. 2007;306:847–859. doi: 10.1016/j.ydbio.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Graham A, Papalopulu N, Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell. 1989;57:367–378. doi: 10.1016/0092-8674(89)90912-4. [DOI] [PubMed] [Google Scholar]
- Izpisua-Belmonte JC, Tickle C, Dolle P, Wolpert L, Duboule D. Expression of the homeobox Hox-4 genes and the specification of position in chick wing development. Nature. 1991;350:585–589. doi: 10.1038/350585a0. [DOI] [PubMed] [Google Scholar]
- Kessel M, Gruss P. Homeotic transformations of murine vertebrae and concomitant alteration of Hox codes induced by retinoic acid. Cell. 1991;67:89–104. doi: 10.1016/0092-8674(91)90574-i. [DOI] [PubMed] [Google Scholar]
- Kmita M, Duboule D. Organizing axes in time and space; 25 years of colinear tinkering. Science. 2003;301:331–333. doi: 10.1126/science.1085753. [DOI] [PubMed] [Google Scholar]
- Kmita M, Kondo T, Duboule D. Targeted inversion of a polar silencer within the HoxD complex re-allocates domains of enhancer sharing. Nat Genet. 2000;26:451–454. doi: 10.1038/82593. [DOI] [PubMed] [Google Scholar]
- Kmita M, Fraudeau N, Herault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002;420:145–150. doi: 10.1038/nature01189. [DOI] [PubMed] [Google Scholar]
- Kmita M, Tarchini B, Zakany J, Logan M, Tabin CJ, Duboule D. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435:1113–1116. doi: 10.1038/nature03648. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lu Y, Goldenberg I, Bei L, Andrejic J, Eklund EA. HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with histone deacetylase 2. J Biol Chem. 2003;278:47792–47802. doi: 10.1074/jbc.M305885200. [DOI] [PubMed] [Google Scholar]
- Luke MP, Sui G, Liu H, Shi Y. Yin Yang 1 physically interacts with Hoxa11 and represses Hoxa11-dependent transcription. J Biol Chem. 2006;281:33226–33232. doi: 10.1074/jbc.M606584200. [DOI] [PubMed] [Google Scholar]
- Manzanares M, Bel-Vialar S, Ariza-McNaughton L, Ferretti E, Marshall H, Maconochie MM, Blasi F, Krumlauf R. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involve auto- and cross-regulatory mechanisms. Development. 2001;128:3595–3607. doi: 10.1242/dev.128.18.3595. [DOI] [PubMed] [Google Scholar]
- Metscher BD, Takahashi K, Crow K, Amemiya C, Nonaka DF, Wagner GP. Expression of Hoxa-11 and Hoxa-13 in the pectoral fin of a basal ray-finned fish, Polyodon spathula: implications for the origin of tetrapod limbs. Evol Dev. 2005;7:186–195. doi: 10.1111/j.1525-142X.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- Montavon T, Duboule D. Chromatin organization and global regulation of Hox gene clusters. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120367. doi: 10.1098/rstb.2012.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Le Garrec JF, Kerszberg M, Duboule D. Modeling Hox gene regulation in digits: reverse collinearity and the molecular origin of thumbness. Genes Dev. 2008;22:346–359. doi: 10.1101/gad.1631708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–919. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, Gonzales E, Tessarollo L, Parada LF, Tabin C. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Duboule D. Chromatin architectures and hox gene collinearity. Curr Top Dev Biol. 2013;104:113–148. doi: 10.1016/B978-0-12-416027-9.00004-8. [DOI] [PubMed] [Google Scholar]
- Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Post LC, Innis JW. Altered Hox expression and increased cell death distinguish Hypodactyly from Hoxa13 null mice. Int J Dev Biol. 1999;43:287–294. [PubMed] [Google Scholar]
- Probst S, Kraemer C, Demougin P, Sheth R, Martin GR, Shiratori H, Hamada H, Iber D, Zeller R, Zuniga A. SHH propagates distal limb bud development by enhancing CYP26B1-mediated retinoic acid clearance via AER-FGF signalling. Development. 2011;138:1913–1923. doi: 10.1242/dev.063966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosello-Diez A, Ros MA, Torres M. Diffusible signals, not autonomous mechanisms, determine the main proximodistal limb subdivision. Science. 2011;332:1086–1088. doi: 10.1126/science.1199489. [DOI] [PubMed] [Google Scholar]
- Schneider I, Shubin NH. The origin of the tetrapod limb: from expeditions to enhancers. Trends Genet. 2013;29:419–426. doi: 10.1016/j.tig.2013.01.012. [DOI] [PubMed] [Google Scholar]
- Shen WF, Krishnan K, Lawrence HJ, Largman C. The HOX homeodomain proteins block CBP histone acetyl-transferase activity. Mol Cell Biol. 2001;21:7509–7522. doi: 10.1128/MCB.21.21.7509-7522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth R, Bastida MF, Ros M. Hoxd and Gli3 interactions modulate digit number in the amniote limb. Dev Biol. 2007;310:430–441. doi: 10.1016/j.ydbio.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Sheth R, Marcon L, Bastida MF, Junco M, Quintana L, Dahn R, Kmita M, Sharpe J, Ros MA. Hox genes regulate digit patterning by controlling the wavelength of a Turing-type mechanism. Science. 2012;338:1476–1480. doi: 10.1126/science.1226804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth R, Gregoire D, Dumouchel A, Scotti M, Pham JM, Nemec S, Bastida MF, Ros MA, Kmita M. Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth. Development. 2013;140:2130–2138. doi: 10.1242/dev.089409. [DOI] [PubMed] [Google Scholar]
- Sordino P, van der Hoeven F, Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature. 1995;375:678–681. doi: 10.1038/375678a0. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- Studer M, Gavalas A, Marshall H, Ariza-McNaughton L, Rijli FM, Chambon P, Krumlauf R. Genetic interactions between Hoxa1 and Hoxb1 reveal new roles in regulation of early hindbrain patterning. Development. 1998;125:1025–1036. doi: 10.1242/dev.125.6.1025. [DOI] [PubMed] [Google Scholar]
- Tabin C, Wolpert L. Rethinking the proximodistal axis of the vertebrate limb in the molecular era. Genes Dev. 2007;21:1433–1442. doi: 10.1101/gad.1547407. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Tschopp P, Duboule D. A regulatory ‘landscape effect’ over the HoxD cluster. Dev Biol. 2011;351:288–296. doi: 10.1016/j.ydbio.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- Woltering JM, Duboule D. The origin of digits: expression patterns versus regulatory mechanisms. Dev Cell. 2010;18:526–532. doi: 10.1016/j.devcel.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Yokouchi Y, Nakazato S, Yamamoto M, Goto Y, Kameda T, Iba H, Kuroiwa A. Misexpression of Hoxa-13 induces cartilage homeotic transformation and changes cell adhesiveness in chick limb buds. Genes Dev. 1995;9:2509–2522. doi: 10.1101/gad.9.20.2509. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature. 1996;384:69–71. doi: 10.1038/384069a0. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Zakany J, Fromental-Ramain C, Warot X, Duboule D. Regulation of number and size of digits by posterior Hox genes: a dose-dependent mechanism with potential evolutionary implications. Proc Natl Acad Sci U S A. 1997;94:13695–13700. doi: 10.1073/pnas.94.25.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakany J, Kmita M, Duboule D. A dual role for Hox genes in limb anterior-posterior asymmetry. Science. 2004;304:1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]