Fig 9. One model to explain the constrained relationship between MutSγ, SIC/Zip1-associated JMs, and MutLγ proteins and how K. l. Zip1 might bypass the constraint.
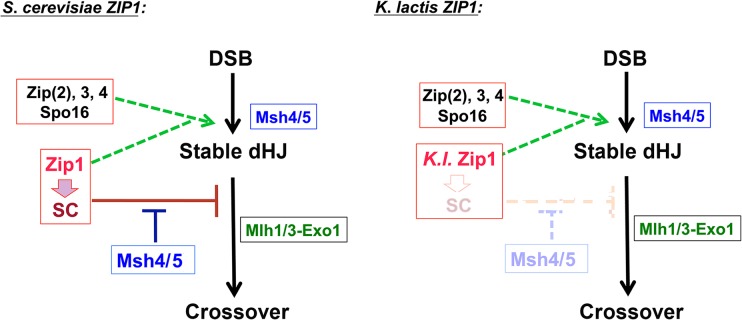
Cartoons depict potential pathways for SC protein-MutSγ-MutLγ associated crossovers in budding yeast. At left is depicted S. cerevisiae expressing S. c. ZIP1, while at right is depicted S. cerevisiae expressing K. l. ZIP1. In these models, Zip1 maintains a pro-crossover function along with other SIC proteins and Msh4/Msh5, upstream of stabilized dHJs. Both S. c. Zip1 and K. l. Zip1 collaborate with other SIC proteins and Msh4/Msh5 to promote the establishment of stable JMs. In this model, S. c. Zip1 also has an anti-crossover activity that Msh4/Msh5 normally counters (left cartoon). This anti-crossover activity might be an intrinsic feature of the Zip1 protein or perhaps is an activity coming from SC formation. An antagonistic relationship between Msh4/Msh5 and S. c. Zip1 constrains Zip1-mediated, MutLγ-associated resolvase activity to act exclusively on MutSγ-associated recombination intermediates. In the context of S. cerevisiae cells expressing K. l. ZIP1 (right cartoon), K. l. Zip1 retains the pro-crossover activity but lacks the anti-crossover activity of S. c. Zip1, rendering MutSγ dispensable for MutLγ-dependent crossover recombination. Since K. l. Zip1 uncouples Mlh3-mediated crossover formation from both SC assembly and from a reliance on Msh4, we are drawn to the possibility that SC assembly normally constrains Zip1 crossovers–ensuring that they occur successfully only on Msh4-associated intermediates. We emphasize that the anti-crossover aspects of Zip1 activity could either be an action that destabilizes JM structures or one that prevents the accessibility of JM structures to MutLγ and other resolvase proteins.
