Abstract
Background
Immunoglobulin γ marker (GM) and κ marker (KM) allotypes, hereditary antigenic determinants of γ and κ chains, respectively, have been shown to be associated with immunity to a variety of self and nonself antigens, but their possible contribution to immunity to the tumor-associated antigens epidermal growth factor receptor (EGFR) and EGFR variant (v)III has not been evaluated. The aim of the present investigation was to determine whether the interindividual variation in endogenous antibody responsiveness to EGFR and EGFRvIII is associated with particular GM, KM, and Fcγ receptor (FcγR) genotypes and whether antibody levels were associated with the overall survival of patients with glioblastoma.
Methods
A total of 126 Caucasian participants with glioblastoma were genotyped for several GM, KM, and FcγR alleles and characterized for IgG antibodies to EGFR and EGFRvIII antigens.
Results
The anti-EGFR antibody levels associated with GM 3/3 homozygotes and GM 3/17 heterozygotes were similar (15.9 vs 16.4 arbitrary units [AU]/µL) and significantly lower than those associated with GM 17/17 homozygotes (19.6 AU/µL; nominal P = .007). Participants homozygous for the GM 21 allele also had significantly higher levels of anti-EGFR antibodies than GM 5/5 homozygotes and GM 5/21 heterozygotes (20.1 vs 16.0 and 16.3 AU/µL; nominal P = .005). Similar associations were found with immune responsiveness to EGFRvIII. Higher anti-EGFR and anti-EGFRvIII antibody levels were associated with enhanced overall survival (16 vs 11 mo, nominal P = .038 and 20 vs 11 mo, nominal P = .004, respectively).
Conclusions
GM allotypes contribute to humoral immunity to EGFR in glioblastoma.
Keywords: antibody, EGFR, EGFRvIII, glioblastoma, GM allotypes
The tumor-associated antigen epidermal growth factor receptor (EGFR) is overexpressed in 40%–60% of patients with malignant gliomas, the most common type of primary brain tumor, which is mostly incurable. EGFR amplification is frequently accompanied by an intragenic rearrangement that produces EGFR variant (v)III, which is tumor specific. Amplification and overexpression of both EGFR and EGFRvIII has been shown to be associated with worse prognosis in glioma patients.1 These observations have made EGFR an attractive target for both active (vaccine) and passive (antibody) immunotherapies against gliomas.2,3 Identification and understanding of the putative host genetic factors that might influence the magnitude of naturally occurring immune responses to EGFR and EGFRvIII is an important prerequisite to successfully designing vaccines and therapeutic antibodies against gliomas. This knowledge would also be important for a proper evaluation of vaccine efficacy trials. Thus, some people could be naturally high responders to EGFR and EGFRvIII, while others could be low responders. A lack of understanding of the host genetic factors involved in EGFR/EGFRvIII immunity hinders effective immunological intervention in glioblastoma and confounds the evaluation of ongoing vaccine efficacy trials.
Immunoglobulin γ marker (GM) and κ marker (KM) allotypes, hereditary antigenic determinants of γ and κ chains, respectively,4,5 have been shown to be associated with immune responsiveness to a variety of antigens—infectious agents, vaccines, autoantigens, including some tumor-associated antigens5–11—but their possible contribution to immunity to EGFR and EGFRvIII has not been evaluated. The importance of Ig genes and humoral immunity in the pathogenesis of solid tumors has been underscored by a recent comprehensive analysis of human gene expression.12 This analysis identified the Ig κ constant (IGKC) gene as a strong prognostic marker in human solid tumors, providing a compelling rationale for investigating the role of KM alleles, genetic variants of IGKC, in the immunopathogenesis of these tumors. It is known that Fcγ receptor (FcγR)–mediated uptake of antigen-antibody complexes can enhance antigen presentation, which provides a good rationale for investigating the role of FcγR genotypes in humoral immunity to EGFR and EGFRvIII. Thus, different FcγR genotypes (of antigen presenting cells) could differentially influence the uptake of IgG opsonized EGFR antigens for presentation to helper T cells, resulting in antigen-specific B-cell activation. FcγR genotypes have been shown to be associated with the magnitude of humoral immunity to some tumor-associated antigens.13
The aim of the present investigation was to determine whether the magnitude of antibody responsiveness to EGFR and EGFRvIII was associated with particular GM, KM, and FcγR genotypes and whether antibody levels were associated with the overall survival of patients with glioblastoma multiforme (GBM). We found that antibody responsiveness to both EGFR and EGFRvIII was associated with particular GM alleles, and higher antibody levels correlated with longer survival.
Methods
Experimental Design
A case-only experimental design was self-controlled. Subjects with a particular genotype were controls for those who lacked this genotype, and vice versa.
Human Participants
The study population comprised a subset of unrelated case participants in the Upper Midwest Health Study, a large population-based, case-control study that evaluated associations between gliomas and environmental and occupational exposures among adults (ages 18–80) residing at diagnosis/selection in nonmetropolitan counties in 4 upper Midwestern states (Iowa, Michigan, Minnesota, Wisconsin).14 Cases were diagnosed between January 1, 1995 and January 31, 1997. Some 798 eligible ascertained cases and 1175 eligible controls provided informed consent. Of 798 cases, 472 fulfilled the World Health Organization criteria of GBM (grade IV glioma). However, most of the GBM participants had died or were too debilitated to donate blood by the time they were ascertained to us. Specimens from 126 Caucasian GBM cases (73 male, 53 female, age range 18–76) were available for the present study.
Donated blood specimens were shipped to the National Institute for Occupational Safety and Health (NIOSH) Cincinnati Laboratory to arrive the morning following the blood draw. Plasma samples were prepared from whole blood using Accuspin System Histopaque-1077 methods (Sigma-Aldrich) and were maintained in a −80°C freezer. Blood and plasma specimens were shipped to the Medical University of South Carolina on dry ice. The study protocol was approved by the institutional review boards of NIOSH and the Medical University of South Carolina.
Gamma Marker and Kappa Marker Genotyping
DNA for genotyping was isolated from peripheral blood using a standard protocol (Qiagen Kit method). For the determination of IGHG1 alleles GM 3 and 17 (arginine to lysine, a G-to-A substitution in the CH1 region of the γ1 gene), we used a predesigned TaqMan genotyping assay from Applied Biosystems. The probe specific to the GM 3 allele was labeled with the fluorescent dye FAM at the 5′ end and with nonfluorescent quencher at the 3′ end. The probe specific to the GM 17 allele was labeled with the fluorescent dye VIC at the 5′ end and with nonfluorescent quencher at the 3′ end.
GM 23—valine to methionine, a G-to-A substitution in the CH2 region of the IGHG2 gene—was determined by a nested PCR–restriction fragment length polymorphism (RFLP) method. In brief, a 915-bp region of the IGHG2 gene that incorporates the sites for the allelic substitutions was amplified as described by Brusco et al,15 using the following primers:
5′ AAATGTTGTGTCGAGTGCCC 3′ and 5′ GGCTTGCCGGCCGTGGCAC 3′. A 197-bp segment was further amplified from this 915-bp fragment using the following primers:
5′ GCACCACCTGTGGCAGGACC 3′ and 5′ TTGAACTGCTCCTCCCGTGG 3′. After digestion of the amplified product with the restriction enzyme NlaIII, the following products corresponding to the 3 genotypes were obtained:
GM 23+ 90 bp, 63 bp, 44 bp
GM 23− 134 bp, 63 bp
GM 23+ 23–134 bp, 90 bp, 63 bp, 44 bp
For the determination of GM 5 and 21 alleles, the IGHG3 gene containing the allelic sites was amplified16 using the following primers:
5′ ACCCAAGGATACCCTTATGATT 3′ and 5′ GAGGCTCTTCTGCGTGAAGC 3′. The amplified product (685 bp) was digested with the restriction enzyme MspA1I. The resulting products corresponding to the 3 genotypes were as follows:
GM 21 327 bp, 295 bp, 63 bp
GM 5 171 bp, 158 bp, 156 bp, 137 bp, 63 bp
GM 5 21 327 bp, 295 bp, 171 bp, 158 bp, 156 bp, 137 bp, 63 bp
Three alleles—KM 1, KM 1,2, and KM 3—segregate at the KM locus in IGKC. The KM 1 allele, without KM 2, is rare; 98% of the individuals positive for KM 1 are also positive for KM 2. Thus, positivity for KM 1 includes both KM 1 and KM 1,2 alleles. KM genotyping was done by a previously described PCR-RFLP method.17
With every experiment, we included known positive and negative controls for the allele being typed. In our experience, various genotyping methods give identical results. This has been formally established in the course of an investigation on the etiology of sarcoidosis, the ACCESS multicenter study coordinated by an independent organization.18,19 For the markers investigated, a complete concordance has been found between the markers determined serologically and those determined at the DNA level, confirmed by independent investigators.17
FcγR Genotyping
The activating receptors FcγRIIa and FcγRIIIa are genetically polymorphic: a change in the nucleotide at position 497 of the FCGR2A gene from A to G results in a change of the amino acid histidine to arginine (H/R131); a change in the nucleotide at position 559 of the FCGR3A gene from T to G results in phenylalanine to valine substitution (F/V158). The FCGR2A alleles were determined by a previously described PCR-RFLP method.20 The FCGR3A alleles were determined by the TaqMan single nucleotide polymorphism genotyping assay, using reagents supplied by Applied Biosystems, following the manufacturer's protocols.
Measurement of Antibodies to EGFR and EGFRvIII
Samples of recombinant human EGFR (Sino Biological) or EGFRvIII (GeneScript USA) were coated in 96-well flat-bottomed plates (20 μg/mL, 50 μL each) at 4°C overnight in 15 mM carbonate-bicarbonate buffer (pH 9.6) as modified after Gupta et al.21 The plates were washed 3 times with phosphate buffered saline containing 0.25% Tween-20 (PBS/T). The plates were blocked with 0.89% bovine serum albumin in PBS/T (blocking buffer) for 1.5 h at room temperature. After washing 3 times with PBS/T, 50 μL of diluted plasma (1:900) prepared in blocking buffer was added to each well in duplicates. Plates were incubated for 60 min at 37°C, the contents were discarded, and the wells were washed 5 times with PBS/T. Horseradish peroxidase–conjugated goat anti-human IgG (γ chain), diluted 1:6000 in blocking buffer, was added. After 30-min incubation at 37°C, wells were washed 5 times with PBS/T, and we added hydrogen peroxide along with TMB (3,3′,5,5′-tetramethylbenzidine; Sigma-Aldrich), 50 μL/well. Plates were incubated for another 15 min at room temperature, and the reaction was stopped by addition of 50 μL of 2N HCl. The plates were read at 450 nm using a microplate reader (BioTek Instruments). The plate-to-plate variation was accounted for by including one positive sample as a reference on every plate. The quantity of anti-EGFR (or anti-EGFRvIII) human IgG detected in respective plasma samples was expressed as absorbance in arbitrary units per microliter (AU/μL) of plasma.
Statistical Analysis
Linear regression models were constructed to test associations between genotypes and anti-EGFR and anti-EGFRvIII IgG antibody responses. Tests of genotype models—2df tests with no assumptions about inheritance models, as well as 1df tests of additive, dominant, and recessive effects of the minor allele—were considered. The phenotypes of interest, anti-EGFR and anti-EGFRvIII IgG antibody levels (ΑU/µL) were transformed (squared) to avoid violating model assumptions. Associations between the prevalence of antibodies and particular GM, KM, and FCGR genotypes and patient survival were assessed using Cox proportional hazards models, adjusting for age. Assumptions of the models were evaluated by exploring Martingale residuals. Log-rank tests were used to evaluate differences in overall survival between participants with high and low antibody levels (dichotomized at the median) using a Kaplan–Meier approach. Statistical significance was defined as P < .05. All reported P-values are 2-sided.
Results
The distribution of KM, GM, and FcγR genotypes among GBM patients in relation to the mean levels of IgG antibodies (AU/µL) to EGFR is given in Table 1. The association between GM 3/17 genotypes and the level of anti-EGFR antibody responses was significant for the genotype model as well as for additive and recessive models, but not for the dominant model of inheritance. The anti-EGFR antibody levels associated with GM 3/3 homozygotes and GM 3/17 heterozygotes were similar (15.9 vs 16.4 AU/µL) and significantly lower than those associated with GM 17/17 homozygotes (19.6 AU/µL; P = .007). The genotypes at the GM 5/21 locus were also associated with anti-EGFR antibody responses for the genotype model as well as for a recessive model of inheritance. Participants homozygous for the GM 21 allele, which is in linkage disequilibrium with GM 17, had significantly higher levels of anti-EGFR antibodies than GM 5/5 homozygotes and GM 5/21 heterozygotes (20.1 vs 16.0 and 16.3 AU/µL; P = .005).
Table 1.
Tests of associations between KM, GM, and FcγR variants and anti-EGFR antibody levels (AU/µL) in patients with glioblastoma
| Loci | Genotype | N | Anti-EGFRvIII Antibody Levels (mean ± SD) | P (genotype) | P (additive) | P (dominant) | P (recessive) |
|---|---|---|---|---|---|---|---|
| KM 1/3 | 3/3 | 104 | 16.7 ± 4.3 | .165 | .141 | .077 | .991 |
| 1/3 | 19 | 14.8 ± 4.0 | |||||
| 1/1 | 3 | 15.8 ± 7.7 | |||||
| GM 3/17 | 3/3 | 60 | 15.9 ± 4.5 | .017 | .021 | .151 | .007 |
| 3/17 | 56 | 16.4 ± 4.1 | |||||
| 17/17 | 10 | 19.6 ± 3.9 | |||||
| GM 5/21 | 5/5 | 63 | 16.0 ± 4.5 | .015 | .057 | .345 | .005 |
| 5/21 | 55 | 16.3 ± 4.0 | |||||
| 21/21 | 8 | 20.1 ± 4.1 | |||||
| GM 23+/− | +/+ | 34 | 15.9 ± 4.4 | .289 | .148 | .392 | .123 |
| +/− | 57 | 16.2 ± 4.5 | |||||
| −/− | 35 | 17.3 ± 4.1 | |||||
| FcγRIIa | R/R | 38 | 16.1 ± 4.3 | .849 | .568 | .609 | .674 |
| R/H | 60 | 16.5 ± 4.5 | |||||
| H/H | 28 | 16.8 ± 4.2 | |||||
| FcγRIIIa | F/F | 49 | 15.6 ± 4.1 | .066 | .358 | .070 | .407 |
| F/V | 62 | 17.3 ± 4.4 | |||||
| V/V | 15 | 15.6 ± 4.4 |
The distribution of KM, GM, and FcγR genotypes among GBM patients in relation to the mean levels of IgG antibodies (AU/µL) to EGFRvIII is given in Table 2. The association between GM 3/17 genotypes and the level of anti-EGFRvIII antibody responses was significant for the recessive model, but not for the genotype, additive, and dominant models of inheritance. GM 17/17 homozygotes had significantly higher anti-EGFRvIII antibody levels than GM 3/17 heterozygotes and GM 3/3 homozygotes (23.1 vs 20.3 and 19.9 AU/µL; P = .043). The association between GM 5/21 genotypes was also significant for the recessive model, but not for the genotype, additive, and dominant models of inheritance. GM 21/21 homozygotes had significantly higher anti-EGFRvIII antibody levels than GM 5/21 heterozygotes and GM 5/5 homozygotes (23.6 vs 20.2 and 20.1 AU/µL; P = .030).
Table 2.
Tests of associations between KM, GM, and FcγR variants and anti-EGFRvIII antibody levels (AU/µL) in patients with glioblastoma
| Loci | Genotype | N | Anti-EGFR Antibody Levels (mean ± SD) | P (genotype) | P (additive) | P (dominant) | P (recessive) |
|---|---|---|---|---|---|---|---|
| KM 1/3 | 3/3 | 104 | 20.7 ± 4.6 | .143 | .084 | .053 | .732 |
| 1/3 | 19 | 18.7 ± 4.2 | |||||
| 1/1 | 3 | 19.0 ± 7.7 | |||||
| GM 3/17 | 3/3 | 60 | 19.9 ± 5.0 | .119 | .118 | .380 | .043 |
| 3/17 | 56 | 20.3 ± 4.3 | |||||
| 17/17 | 10 | 23.1 ± 3.7 | |||||
| GM 5/21 | 5/5 | 63 | 20.1 ± 5.0 | .089 | .231 | .685 | .030 |
| 5/21 | 55 | 20.2 ± 4.2 | |||||
| 21/21 | 8 | 23.6 ± 3.9 | |||||
| GM 23+/− | +/+ | 34 | 19.7 ± 4.7 | .276 | .120 | .283 | .134 |
| +/− | 57 | 20.1 ± 5.0 | |||||
| −/− | 35 | 21.4 ± 3.9 | |||||
| FcγRIIa | R/R | 38 | 20.3 ± 4.7 | .968 | .938 | .946 | .835 |
| R/H | 60 | 20.4 ± 4.8 | |||||
| H/H | 28 | 20.3 ± 4.3 | |||||
| FcγRIIIa | F/F | 49 | 19.8 ± 4.4 | .122 | .848 | .263 | .198 |
| F/V | 62 | 21.1 ± 4.7 | |||||
| V/V | 15 | 18.9 ± 4.9 |
KM and FcγR genotypes were not associated with antibody responsiveness to EGFR or EGFRvIII. Also, there was no significant interactive effect of these genotypes on antibody responsiveness (data not shown).
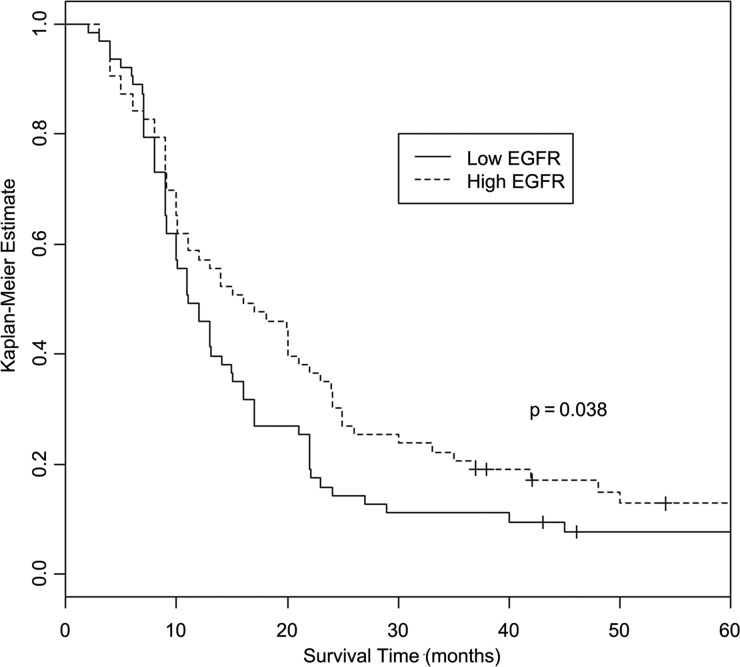
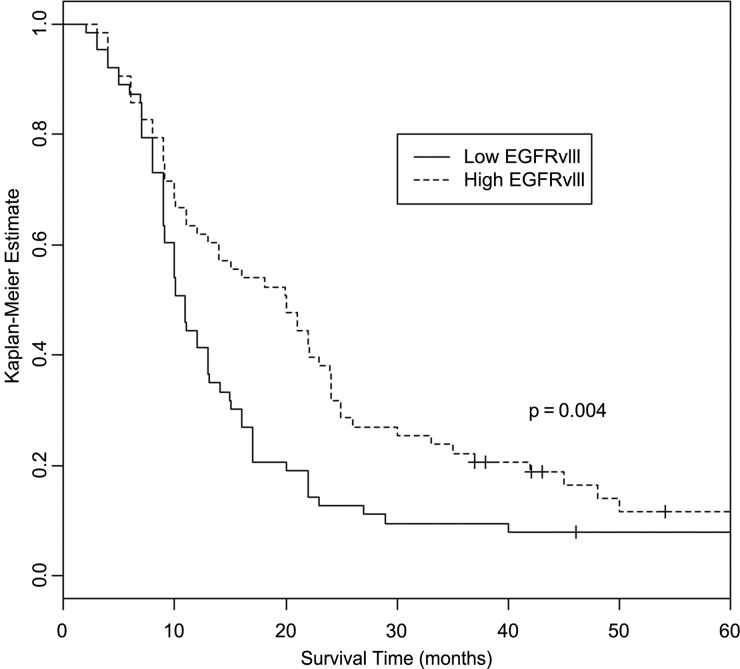
Analyses of association between EGFR antibody levels and overall survival, using Cox proportional hazards models, showed significant increase in survival with increasing antibody levels (hazard ratio = 0.95 [95% CI: 0.91–0.99], P = .027). Analyses of association between EGFRvIII antibody levels and overall survival were not statistically significant (hazard ratio = 0.97 [95% CI: 0.93–1.02], P = .177). Kaplan–Meier curves showed significant differences in overall survival by low/high (dichotomized at the median) EGFR and EGFRvIII antibody levels. For EGFR antibodies, median overall survival for patients with high antibody levels was about 45% longer than those with low antibody levels (16 mo [95% CI: 11–23] vs 11 mo [95% CI: 10–15], P = .038; Fig. 1). For EGFRvIII antibodies, median overall survival for patients with high antibody levels was about 82% longer than those with low antibody levels (20 mo [95% CI: 13–24] vs 11 mo [95% CI: 9–31], P = .004; Fig. 2). These observations suggest that EGFR and EGFRvIII antibodies may influence survival time in a nonlinear fashion. Participants with GM alleles (17 and 21) that were associated with high antibody responsiveness to these tumor-associated antigens survived longer, but the results were not statistically significant (P = .075 and .118, respectively), which could be a reflection of small sample size.
Fig. 1.
Kaplan–Meier curves showing differences in overall survival by low/high EGFR antibody level (dichotomized at the median). Log-rank test shows the association between higher EGFR antibody level and longer survival (P = .038).
Fig. 2.
Kaplan–Meier curves showing differences in overall survival by low/high EGFRvIII antibody level (dichotomized at the median). Log-rank test shows the association between higher EGFRvIII antibody level and longer survival (P = .004).
The nominal P-values are given here with the caveat that they were not adjusted for multiple testing, so that the reader can make an informed judgment. Adjustments for multiple testing are controversial,22 and we believe that instead of performing such adjustment in this work, the best approach would be to test in an independent sample. It is relevant to point out that because of significant linkage disequilibrium within GM and FcγR loci, not all tests performed in this study were independent. Thus, associations at 3 independent loci—GM, KM, and FcγR—were explored. The P-values for the associations between GM genotypes and antibody responsiveness to EGFR, but not to EGFRvIII, would remain significant even after a conservative correction for multiple testing, assuming the recessive model (the most appropriate for the data) of inheritance.
Discussion
The results presented here show that GBM patients who were homozygous for the γ1 determinant GM 17 had higher levels of IgG antibodies to both EGFR and its variant EGFRvIII than those with the other 2 genotypes at this locus. Similar results were obtained for the γ3 determinant GM 21. Although we did not determine the subclasses of EGFR/EGFRvIII IgG antibodies in this study, the majority of the T-cell–dependent antibody responses are known to be IgG1. Thus, GM 17 (expressed on IgG1) might be the primary determinant of high antibody responsiveness to EGFR, and the association of GM 21 might be a result of its almost absolute significant linkage disequilibrium with GM 17 in Caucasians. Significant linkage disequilibrium between GM 17 and the variable region genes involved in anti-EGFR/EGFRvIII antibody responsiveness could explain the observed associations.
Another mechanism underlying the observed association could involve GM determinants being part of the recognition structure for the EGFR epitopes on the membrane-bound IgG (mIgG) of memory B cells. IgG-expressing memory B cells show enhanced response to antigen stimulation compared with cells expressing IgM on their surface.23,24 GM allotype-caused/associated structural changes could influence the magnitude of antibody responsiveness through the antigen processing/presenting pathway. Perhaps B-cell mIgG molecules expressing GM 17 and GM 21 determinants constitute a higher affinity receptor for the EGFR/EGFRvIII epitopes and are more efficient than others in the uptake of these antigens and presenting them to collaborating helper T cells, thus resulting in higher B-cell activation. The magnitude of antibody responsiveness to EGFR is different from that of EGFRvIII. This suggests that the immunogenic epitopes recognized by the endogenous antibodies for the 2 tumor antigens are also different.
The constant-region GM variants could also influence antibody affinity and specificity by causing conformational changes in the variable regions of γ chains. Increasing evidence from experimental organisms supports this contention.25 It is especially noteworthy that amino acid sequence polymorphism in the CH1 domain of the γ1 chain, where GM 17 is located, has been shown to modulate the kinetic competence of antigen binding sites.26 Additionally, GM determinants could influence the expression of idiotypes involved in EGFR/EGFRvIII immunity. Although not yet investigated in humans, both variable and constant regions have been shown to be involved in the formation of certain idiotypic determinants in mice.27 It is also possible that the associations we have observed are due to linkage disequilibrium between these GM alleles and alleles of another locus, as yet unidentified, for humoral immune responsiveness to EGFR/EGFRvIII.
As mentioned before, EGFR/EGFRvIII peptides are being evaluated as therapeutic vaccines in GBM patients.2,3,28 Given the genetically heterogeneous nature of the human population, however, it is unlikely that all vaccinees would respond equally well to a given EGFR/EGFRvIII vaccine regimen. Results from numerous studies of immune responsiveness to vaccines against infectious agents support this contention.29–31 Analogous to the interindividual differences in the naturally occurring endogenous antibody responses to EGFR/EGFRvIII peptides observed in this study, variability in immune responsiveness is likely to occur in response to any administered vaccine regimen as well. This suggests that a personalized approach to immunotherapeutic interventions in patients with GBM might be more effective. Toward this end, the results presented here, if confirmed in an independent study, could aid in identifying participants (GM 17+, GM 21+) who are more likely to benefit from EGFR/EGFRvIII-based vaccines. For participants with the low responder GM genotype (GM 3+, GM 5+), EGFR/EGFRvIII could be fused with appropriate adjuvants, such as heat shock proteins or flagellin, to overcome the GM allotype-associated restriction in humoral immunity. It is relevant to note that antibody responses to certain heat shock proteins as well as to flagellin are also influenced by GM genotypes,32,33 making it conceivable to formulate a fusion EGFR/EGFRvIII–heat shock protein/flagellin vaccine regimen that could potentially generate high antibody responses in the majority of the population.
GBM patients with high levels of antibodies to EGFR and EGFRvIII survived longer than those with low levels of antibodies to these tumor-associated antigens. Likely immunological mechanisms underlying the beneficial effect of these antibodies could involve IgG Fc-mediated effector functions—antibody-dependent cell-mediated cytotoxicity (ADCC), antibody-dependent complement-dependent cytotoxicity (ADCDC), and antibody-dependent cellular phagocytosis. Ligation of the Fc region of IgG antibodies to the FcγRs expressed on effector cells or to the C1q complex triggers these effector functions. The majority of the GM determinants are expressed on the Fc portion of the IgG molecule. We have presented evidence34 for the involvement of GM allotypes in the ADCC of EGFR/human epidermal growth factor receptor (HER)1 and EGFR2/HER2-overexpressing cells from breast cancer and epidermal cancer cell lines (SKBR-3 and A431). Similar studies involving GBM cell lines are warranted.
The complement system also plays an important role in immunosurveillance, and ADCDC could be another mechanism underlying the beneficial effect of endogenous anti-EGFR and anti-EGFRvIII antibodies on the survival of GBM patients. In ADCDC, the C1q complex binds the antibody and triggers the complement cascade. The binding affinity of C1q to the antibody molecules is likely to affect the level of complement-dependent cytotoxicity against tumor cells. It has been known for some time that C1q discriminates between 2 major alleles of IGHG3: it binds slightly better to IgG3 proteins expressing the GM 21 allele than to those expressing the alternative GM 5 allele.35 It follows that antitumor IgG3 antibodies expressing the GM 21 allele in their Fc region would probably be more effective in ADCDC against cancer cells than those expressing the GM 5 allele. Our finding that GBM patients who were homozygous for the GM 21 allele had higher levels of IgG antibodies to both EGFR and its variant EGFRvIII, which, in turn, were associated with better overall survival, is consistent with this observation.
All patients in the present study were Caucasians. In view of the marked racial differences in GM allele frequencies and in the prevalence of GBM, studies in other racial groups are also warranted. In addition to GM alleles, it would be of interest to explore the individual and epistatic role of human leukocyte antigen (HLA) alleles, as virtually all T-cell–dependent antibody responses are HLA restricted, and both HLA and GM alleles (individually and interactively) have been shown to be associated with antibody responsiveness to other T-cell–dependent antigens.33 Mechanisms underlying the epistatic interaction between GM and HLA alleles in humoral immunity to EGFR could involve the recognition of EGFR antigens by the B-cell membrane-bound, allotypically disparate IgG receptors, followed by processing and presentation to the peptide-binding groove of the relevant HLA alleles.
This is the first study of its kind and it needs to be replicated and extended by independent investigations.
Funding
This work was supported in part by the National Institute of Neurological Disorders and Stroke (NS078545) and the National Institute for Occupational Safety and Health. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
Acknowledgment
The assistance of John Clark of NIOSH in assembling the specimens is gratefully acknowledged.
Conflict of interest statement. None declared.
References
- 1.Heimberger AB, Hlatky R, Suki D, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in glioblastoma multiforme patients. Clin Cancer Res. 2005;11(14):1462–1466. [DOI] [PubMed] [Google Scholar]
- 2.Ye F, Gao Q, Cai MJ. Therapeutic targeting of EGFR in malignant gliomas. Expert Opin Ther Targets. 2010;14(3):303–316. [DOI] [PubMed] [Google Scholar]
- 3.Schmittling RJ, Archer GE, Mitchell DA, et al. Detection of humoral response in patients with glioblastoma receiving EGFRvIII-KLH vaccines. J Immunol Methods. 2008;339(1):74–81. [DOI] [PubMed] [Google Scholar]
- 4.Grubb R. Advances in human immunoglobulin allotypes. Exp Clin Immunogenet. 1995;12(3):191–197. [DOI] [PubMed] [Google Scholar]
- 5.Pandey JP, Li Z. The forgotten tale of immunoglobulin allotypes in cancer risk and treatment. Exp Hematol Oncol. 2013;2(3):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey JP, Namboodiri AM, Kistner-Griffin E. IgG and FcγR genotypes and humoral immunity to mucin 1 in prostate cancer. Hum Immunol. 2013;74(8):1030–1033. [DOI] [PubMed] [Google Scholar]
- 7.Pandey JP, Nietert PJ, Mensdorff-Pouilly S, et al. Immunoglobulin allotypes influence antibody responses to mucin 1 in patients with gastric cancer. Cancer Res. 2008;68(11):4442–4446. [DOI] [PubMed] [Google Scholar]
- 8.Pandey JP, Nietert PJ, Klaamas K, et al. A genetic variant of immunoglobulin γ2 is strongly associated with natural immunity to mucin 1 in patients with breast cancer. Cancer Immunol Immunother. 2009;58(12):2025–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey JP, Namboodiri AM, Kurtenkov, et al. Genetic regulation of antibody responses to human epidermal growth factor receptor 2 (HER-2) in breast cancer. Hum Immunol. 2010;71(11):1124–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey JP, Namboodiri AM, Kistner-Griffin E, et al. Racially restricted contribution of immunoglobulin Fcγ and Fcγ receptor genotypes to humoral immunity to human epidermal growth factor receptor 2 in breast cancer. Clin Exp Immunol. 2013;171(3):273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandey JP, Shannon BT, Tsang KY, et al. Heterozygosity at GM loci associated with humoral immunity to osteosarcoma. J Exp Med. 1982;155(4):1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt M, Hellwig B, Hammad S, et al. A comprehensive analysis of human gene expression profiles identifies stromal immunoglobulin kappa C as a compatible prognostic marker in human solid tumors. Clin Cancer Res. 2012;18(9):2695–2704. [DOI] [PubMed] [Google Scholar]
- 13.Pandey JP, Namboodiri AM, Kistner-Griffin E. A genetic variant of FcγRIIIa is strongly associated with humoral immunity to cyclin B1 in African American patients with prostate cancer. Immunogenetics. 2013;65(2):91–96. [DOI] [PubMed] [Google Scholar]
- 14.Ruder AM, Waters MA, Carreon T, et al. The Upper Midwest Health Study: a case-control study of primary intracranial gliomas in farm and rural residents. J Agric Saf Health. 2006;12(4):255–274. [DOI] [PubMed] [Google Scholar]
- 15.Brusco A, de Lange GG, Boccazzi C, et al. Molecular characterization of G2m(n+) and G2m(n−) allotypes. Immunogenetics. 1995;42(5):414–417. [DOI] [PubMed] [Google Scholar]
- 16.Balbín M, Grubb A, de Lange GG, et al. DNA sequences specific for Caucasian G3m(b) and (g) allotypes: allotyping at the genomic level. Immunogenetics. 1994;39(3):187–193. [DOI] [PubMed] [Google Scholar]
- 17.Moxley G, Gibbs RS. Polymerase chain reaction-based genotyping for allotypic markers of immunoglobulin kappa shows allelic association of KM with kappa variable segment. Genomics. 1992;13(1):104–108. [DOI] [PubMed] [Google Scholar]
- 18.ACCESS Research Group. Design of A Case Control Etiologic Study of Sarcoidosis (ACCESS). J Clin Epidemiol. 1999;52(12):1173–1186. [DOI] [PubMed] [Google Scholar]
- 19.Pandey JP, Frederick M, ACCESS Research Group. TNF-α, IL1-β, and immunoglobulin (GM and KM) gene polymorphisms in sarcoidosis. Hum Immunol. 2002;63(6):485–491. [DOI] [PubMed] [Google Scholar]
- 20.Jiang XM, Arepally G, Poncz M, et al. Rapid detection of the Fc gamma RIIA-H/R 131 ligand-binding polymorphism using an allele-specific restriction enzyme digestion (ASRED). J Immunol Methods. 1996;199(1):55–59. [DOI] [PubMed] [Google Scholar]
- 21.Gupta P, Han SY, Holgado-Madruga M, et al. Development of an EGFRvIII specific recombinant antibody. BMC Biotechnol. 2010;10:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perneger TV. What’s wrong with Bonferroni adjustments. BMJ. 1998;316(7139):1236–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin SW, Goodnow CC. Burst-enhancing role of the IgG membrane tail as a molecular determinant of memory. Nat Immunol. 2002;3(2):182–188. [DOI] [PubMed] [Google Scholar]
- 24.Wakabayashi C, Adachi T, Wienands J, et al. A distinct signaling pathway used by the IgG-containing B cell antigen receptor. Science. 2002;298(5602):2392–2395. [DOI] [PubMed] [Google Scholar]
- 25.Casadevall A, Pirofski L-A. A new synthesis for antibody-mediated immunity. Nat Immunol. 2012;13(1):21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres M, Fernandez-Fuentes N, Fiser A, et al. The immunoglobulin heavy chain constant region affects kinetic and thermodynamic parameters of antibody variable region interactions with antigen. J Biol Chem. 2007;282(18):13917–13927. [DOI] [PubMed] [Google Scholar]
- 27.Morahan G, Berek C, Miller JFAP. An idiotypic determinant formed by both immunoglobulin constant and variable regions. Nature. 1983;301(5902):720–722. [DOI] [PubMed] [Google Scholar]
- 28.Foy KC, Wygle RM, Miller MJ, et al. Peptide vaccines and peptidomimetics of EGFR (HER-1) ligand binding domain inhibit cancer cell growth in vitro and in vivo. J Immunol. 2013;191(1):217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pandey JP. Immunoglobulin GM and KM allotypes and vaccine immunity. Vaccine. 2001;19(6):613–617. [DOI] [PubMed] [Google Scholar]
- 30.Ovsyannikova IG, Jacobson RM, Vierkant RA, et al. HLA supertypes and immune responses to measles-mumps-rubella viral vaccine: findings and implications for vaccine design. Vaccine. 2007;25(16):3090–3100. [DOI] [PubMed] [Google Scholar]
- 31.Dhiman N, Ovsyannikova IG, Cunningham JM, et al. Associations between measles vaccine immunity and single-nucleotide polymorphisms in cytokine and cytokine receptor genes. J Infect Dis. 2007;195(1):21–29. [DOI] [PubMed] [Google Scholar]
- 32.Pandey JP, Prohászka Z, Veres A, et al. Epistatic effects of genes encoding immunoglobulin GM allotypes and interleukin-6 on the production of autoantibodies to 60- and 65-kDa heat-shock proteins. Genes Immun. 2004;5(1):68–71. [DOI] [PubMed] [Google Scholar]
- 33.Whittingham S, Mathews JD, Schanfield MS, et al. Interactive effect of GM allotypes and HLA-B locus antigens on the human antibody response to a bacterial antigen. Clin Exp Immunol. 1980;40(1):8–15. [PMC free article] [PubMed] [Google Scholar]
- 34.Namboodiri AM, Pandey JP. Differential inhibition of trastuzumab and cetuximab induced cytotoxicity of cancer cells by IgG1 expressing different GM allotypes. Clin Exp Immunol. 2011;166(3):361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brüggemann M, Williams GT, Bindon CI, et al. Comparison of the effector functions of human immunoglobulins using a matched set of chimeric antibodies. J Exp Med. 1987;166(5):1351–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]