Figure 5. Genome partition.
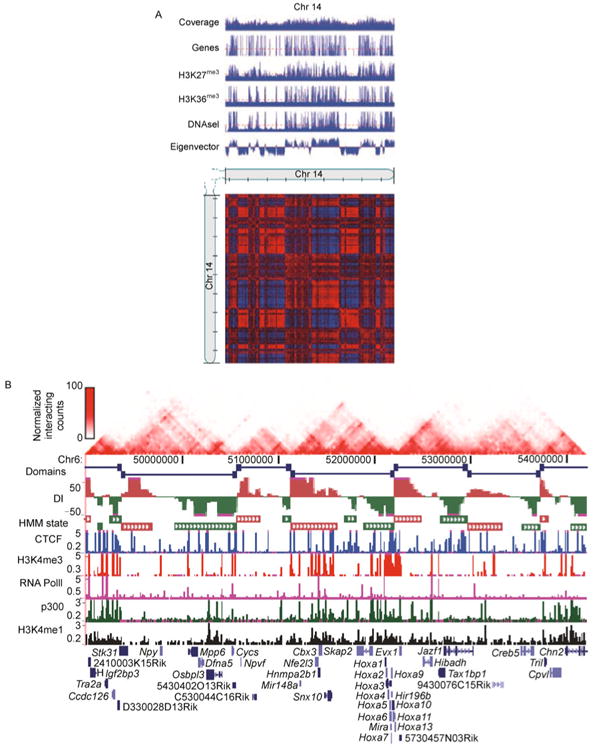
(A) Compartment labels are associated with different genetic and epigenetic markers (direct copy of Ref. [34], Figure 3G). We use chromosome 14 in human GM06990 Hi-C dataset [34] as an example. The Hi-C contact map and genetic and epigenetic markers are at 100 KB resolution. A PCA based approach was applied to the normalized Hi-C contact map to obtain eigenvectors. The compartment A is defined as those 100 KB bins with positive eigenvectors, and the compartment B is defined as those 100 KB bins with negative eigenvectors. Compartment A is associated with gene rich, actively transcribed regions and compartment B is associated with gene poor, repressively transcribed regions. (B) Topological domains appear to serve as units of genomic organization and perhaps function (direct copy of Ref. [45], Figure 1a). We use chromosome 6 in mouse embryonic stem cell Hi-C dataset [45] as an example. The Hi-C contact map and genetic and epigenetic markers are at 40 KB resolution. A hidden Markov model based approach was applied to the normalized Hi-C contact map to identify topological domain boundaries. Overlap of topological domains and other genetic and epigenetic features revealed that the domain boundary regions are enriched for the insulator binding protein CTCF and house-keeping genes.
