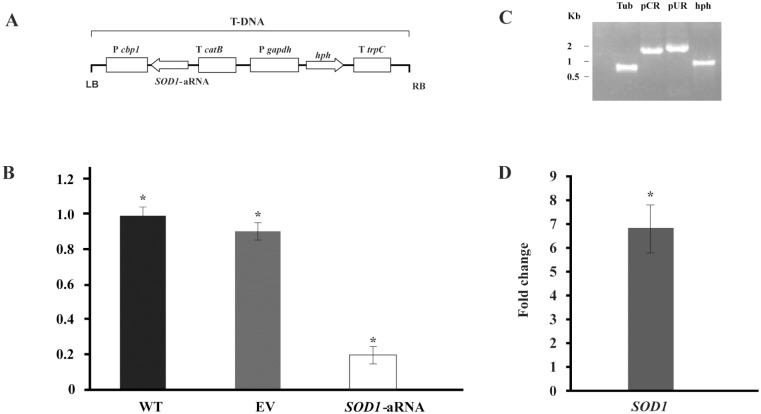
Fig 6. Gene silencing of SOD1 in P. brasiliensis.
(A) Transfer DNA (T-DNA) inserted into the genome of P. brasiliensis yeast cells via ATMT in order to silence the SOD1 gene. The antisense oligonucleotide was directed to exon 3 (black box) that amplify a length of 85 bp. This AS oligonucleotide was placed under the control of the calcium binding protein (CBP-1) with a terminator (CAT-B); the plasmid contained hygromycin B phosphotransferase (HPH) under the control of glyceraldehyde 3-phosphate of Aspergillus nidulans (PGPDA) with a terminator (TTRCP). (B) PbSOD1 gene expression levels obtained by RT-qPCR. The measurement was normalized with the housekeeping gene alpha-tubulin in WT, EV and SOD1-aRNA yeast cells growing in the exponential phase. Mitotic stability was confirmed by sub-culturing P. brasiliensis SOD1-aRNA yeast cells and testing for low expression levels in this isolate after successive sub-cultures. (C) Validation by PCR of the presence and integration of the Transfer DNA (T-DNA) into the genome of P. brasiliensis transformant. The genomic DNA from the SOD1-aRNA isolate was tested by PCR using specific primers for the alpha-tubulin gene TUB (Tub, lane 3), for the transformation constructs pCR35 (pCR, lane 4) and pUR5750 (pUR, lane 5) and for the hygromycin resistance gene (hph, lane 6). (D) SOD1 expression profile in P. brasiliensis after exposure to TSC-C. RNA was extracted after 8 h of exposure of yeast cells to TSC-C. Changes in gene expression levels were calculated by the relative standard curve method using the non-treated control samples to calibrate.