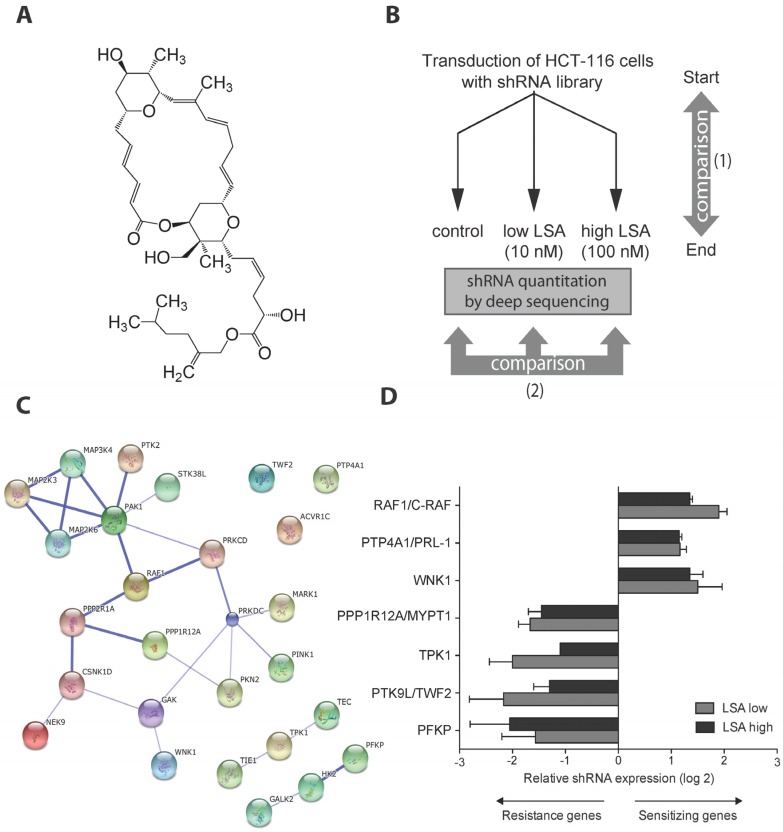
Figure 1.
shRNA screen for genetic determinants of lasonolide A (LSA) sensitivity. (A) Chemical structure of LSA; (B) Scheme of the shRNA screen aiming to identify genes that modulate LSA sensitivity. A pooled retroviral shRNA library directed against kinases and phosphatases was transduced into HCT-116 colorectal cancer cells. Cells were cultured in the absence (control) or presence of LSA at two concentrations, 10 nM for 24 days or 100 nM for 14 days, respectively. Library compositions in starting and end samples were deconvoluted by deep sequencing to identify shRNAs that selectively dropped out or enriched in the LSA-treated samples; (C) Top candidates are enriched in known protein-protein interactions as determined by STRING; (D) shRNAs that enriched and dropped out selectively in the LSA treated samples. Results are expressed as the difference between the log2 ratio of (LSA/start) and the log2 ratio of (control/start). LSA low and high indicates the 10 nM and 100 nM treatment conditions, respectively. Genes whose shRNAs enriched with LSA treatment are potentially associated with LSA sensitivity, while genes whose shRNAs depleted with LSA treatment are potentially associated with resistance.