Abstract
Adjacent alternative 3′ splice sites, those separated by ≤18 nucleotides, provide a unique problem in the study of alternative splicing regulation; there is overlap of the cis-elements that define the adjacent sites. Identification of the intron's 3′ end depends upon sequence elements that define the branchpoint, polypyrimidine tract, and terminal AG dinucleotide. Starting with RNA-seq data from germline-enriched and somatic cell-enriched Caenorhabditis elegans samples, we identify hundreds of introns with adjacent alternative 3′ splice sites. We identify 203 events that undergo tissue-specific alternative splicing. For these, the regulation is monodirectional, with somatic cells preferring to splice at the distal 3′ splice site (furthest from the 5′ end of the intron) and germline cells showing a distinct shift toward usage of the adjacent proximal 3′ splice site (closer to the 5′ end of the intron). Splicing patterns in somatic cells follow C. elegans consensus rules of 3′ splice site definition; a short stretch of pyrimidines preceding an AG dinucleotide. Splicing in germline cells occurs at proximal 3′ splice sites that lack a preceding polypyrimidine tract, and in three instances the germline-specific site lacks the AG dinucleotide. We provide evidence that use of germline-specific proximal 3′ splice sites is conserved across Caenorhabditis species. We propose that there are differences between germline and somatic cells in the way that the basal splicing machinery functions to determine the intron terminus.
Alternative splicing is a highly regulated process by which a cell can produce multiple messenger RNAs, potentially encoding multiple proteins, from a common precursor transcript. The de novo assembly of a spliceosome on each intron of a pre-mRNA transcript requires cis-elements within the intron that are recognized as signals marking the beginning, end, and branchpoint (Ares and Weiser 1995). A cassette exon is a form of alternative splicing in which an exon is either included or skipped in the mature mRNA. Conserved enhancer or silencer elements within the exon or the surrounding introns interact with an array of constitutive and tissue-specific trans-factors that promote or inhibit assembly of a functional spliceosome (Wang and Burge 2008). The use of alternative 3′ or 5′ splice sites modifies gene expression by including or skipping coding sequences at the ends of exons. Many examples of adjacent alternative 3′ splice sites, defined as being separated by 18 nucleotides (nt) or less, have been observed. In many species, a form of alternative 3′ splice site usage has been identified in which the alternative splicing acceptors are only 3 nt apart. Except for the rare example of AC dinucleotides observed for substrates of the minor spliceosome, introns end with AG dinucleotides. Alternative 3′ splice sites separated by only 3 nt are referred to as NAGNAGs, as the end of the intron consists of two AG splice acceptors separated by 3 nt. A recent report provided strong evidence that NAGNAG alternative splicing can be regulated in a tissue-specific manner in mammals (Bradley et al. 2012). The close proximity of these sites to each other makes the influence of enhancer or silencer elements on splicing to one site or the other, as with cassette exons, unlikely. Therefore, NAGNAGs provide an interesting model in which to understand more about the regulation of alternative splicing and, more specifically, the mechanisms by which a 3′ splice site is chosen.
3′ splice sites are determined by a combinatorial code that consists of nucleotides found within the site itself as well as within the intron that precedes it. Typical 3′ splice sites consist of an AG dinucleotide and a stretch of upstream, intronic pyrimidines that bind to U2AF35 and U2AF65, respectively (Merendino et al. 1999; Zorio and Blumenthal 1999; Wu et al. 2011). This complex associates with SF1/BBP, which binds to a conserved sequence surrounding an intronic adenosine nucleotide as the branchpoint for that intron (Berglund et al. 1997). Together, SF1 and the U2AF35/U2AF65 complex recruit the U2 snRNP to the branchpoint region and promote spliceosome assembly (Zhang et al. 1992; Berglund et al. 1998). The scanning model proposes that the 3′ splice site is determined by identification of the first AG dinucleotide downstream from the branchpoint, irrespective of sequence context or distance from the branchpoint (Smith et al. 1989). A modified scanning model allows for some variability in 3′ splice site choice in a small range of nucleotides within the reach of the spliceosome (Smith et al. 1993) but still suggests that the mechanism of 3′ splice site choice, even at NAGNAGs, is stochastic and common to every tissue. Refuting this model, a global analysis of gene expression in 16 human and eight mouse tissues revealed that splicing patterns at individual NAGNAGs are tissue-specific, regulated, and conserved (Bradley et al. 2012). This work highlighted splicing patterns at adjacent 3′ splice sites that suggest a role for the polypyrimidine tract, the location of the branchpoint, and the identity of the N within each NAGNAG in designating a site as the end of an intron (Bradley et al. 2012). It is still unclear, though, if these contributions apply equally to splicing patterns at adjacent 3′ splice sites in every cell type and to what extent these factors influence splice site choice in other organisms.
C. elegans is an excellent model organism in which to study developmental and tissue-specific alternative splicing. The developmental lineages of all 959 somatic cells have been traced (Sulston and Horvitz 1977), and the C. elegans genome was the first among animals to be fully sequenced (The C. elegans Sequencing Consortium 1998). Intergenic regions as well as typical introns are relatively small (Spieth and Lawson 2006). Roughly 25% of protein-coding genes in C. elegans have multiple isoform annotations, with each gene producing 2.2 isoforms on average (Ramani et al. 2011). In addition, many C. elegans trans-acting alternative splicing factors and their targeted cis-elements are conserved in mammals, making these worms an excellent model organism to study human diseases related to splicing factors (Kabat et al. 2006; Zahler 2012; Barberan-Soler and Ragle 2013). The 5′ splice site consensus sequence in C. elegans is similar to other eukaryotes. In nematodes, introns end in AG and are preceded by a short polypyrimidine tract yielding the consensus sequence YYYNAG/R (Blumenthal and Steward 1997). This short polypyrimidine tract differs from other animals in that it is much shorter and located closer to the AG dinucleotide. The YYYNAG/R consensus sequence is a direct binding site for the C. elegans U2AF subunits UAF-1 and UAF-2 (Zorio and Blumenthal 1999; Hollins et al. 2005). The phenomenon of NAGNAG alternative 3′ splice sites separated by 3 nt is rare in C. elegans, presumably due to the strong nematode 3′ splice site consensus with its adjacent pyrimidines precluding a 3-nt separation of two AG dinucleotide splice acceptors. A recent example of tissue-specific use of adjacent alternative 3′ splice sites separated by 9 nt has been uncovered in the C. elegans let-363 gene (Barberan-Soler et al. 2014). The splice acceptor site closer to the 5′ end of the intron (proximal) is favored in the germline, while the site further from the 5′ end (distal) is preferred in somatic tissue. Regulation of this splicing in the male germline was shown to be dependent on antisense transcription of a gene located within an intron of let-363, which is controlled by the piRNA pathway. The extent to which adjacent 3′ splice sites in C. elegans may be utilized, conserved, or regulated on a tissue-specific level is unknown. Further study of these adjacent 3′ splice sites may contribute to our understanding of the mechanisms of 3′ splice site choice by the spliceosome and spliceosome-associated factors. In this study, we uncover and characterize tissue-specific use of 203 alternative adjacent 3′ splice sites. In all cases, there is tissue-specific usage of the proximal splice site in the germline relative to somatic tissue.
Results
Alternative splicing regulation of top-1 and establishing a method for detecting germline-specific splicing
We set out to study the regulation of alternative splicing in C. elegans in a specific tissue, the germline, which develops fully during the transition to adulthood. Previous studies in C. elegans have focused on the developmental timing and regulation of alternative splicing (Kuroyanagi et al. 2000; Barberan-Soler and Zahler 2008; Barberan-Soler et al. 2009; Ramani et al. 2011). For example, top-1 alternative cassette exon splicing patterns change during development (Lee et al. 1998), and RNAi knockdown of the developmentally regulated splicing factor gene hrpf-1 leads to a change in cassette exon inclusion in top-1 isoforms (Barberan-Soler and Zahler 2008). A recent study used immunostaining with antipeptide antibodies specific to the alternative cassette exon of top-1 to demonstrate detection of the skipping isoform of top-1 in almost all cells and the inclusion isoform of top-1 in neuronal cells, excretory cells, and the germline (Cha et al. 2012). We set out to test whether alternative splicing of top-1 pre-mRNA changes with the onset of development of the mature gonad, which corresponds with developmental expansion of the germline. Total RNA was extracted from wild-type L3 and L4 larval stage worms prior to the establishment of a mature germline, as well as wild-type young adult worms following establishment of the mature germline. Reverse transcription with random hexamers followed by polymerase chain reaction (RT-PCR) with primers that anneal to sequences in exons that flank the alternative cassette exon reveals a shift to inclusion of the top-1 cassette exon upon development of a mature germline (Fig. 1A). Because RNA was extracted from whole worms, inclusion of the top-1 alternative exon in mature adults could be a result of overall changes in alternative splicing in the worm as it ages or, more specifically, to growth and maturity of a new organ, the gonad, containing an expanded germline.
Figure 1.
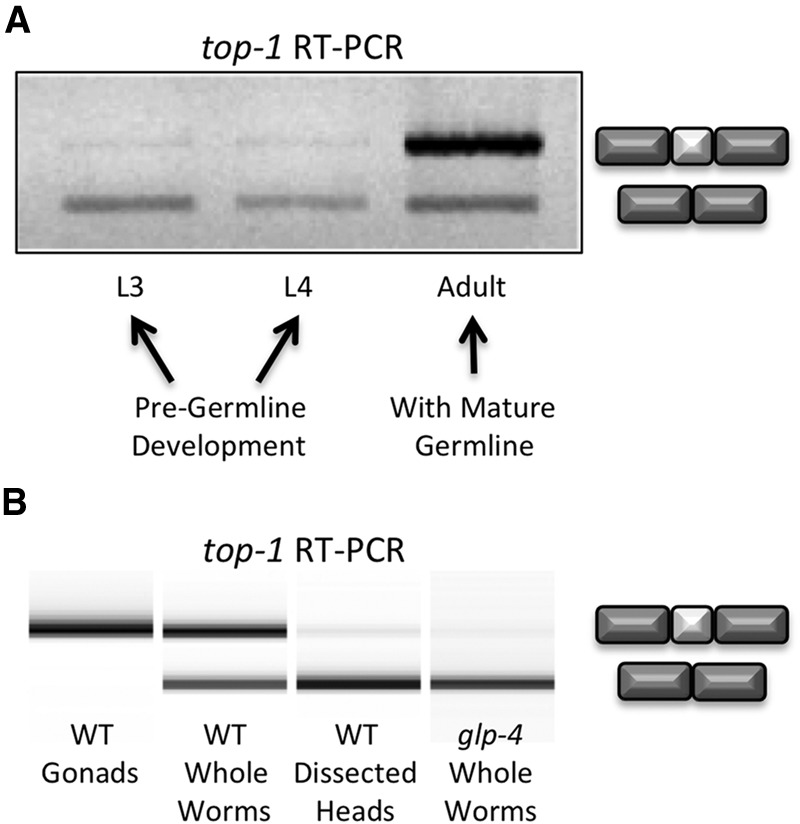
Developmental and tissue-specific changes in alternative splicing of the top-1 gene. (A) Exon inclusion of top-1 alternative cassette exon occurs with the onset of germline development. A 1% agarose gel of reverse-transcription-polymerase chain reaction (RT-PCR) products of RNA extracted from handpicked C. elegans whole worms at indicated developmental stages. Exon inclusion and skipping products are indicated at right. (B) A 2100 Bioanalyzer image of top-1 RT-PCR products of RNA extracted from L4+1 day C. elegans dissected wild-type gonads, adult whole worms, and dissected heads, along with glp-4(bn2ts) mutant whole worms that fail to develop a germline.
To address whether changes in top-1 alternative splicing in adults were due to tissue-specific splicing in the germline, young adult worms 20–24 h past L4 were dissected to isolate gonads and heads. Total RNA was extracted from these tissues as well as from whole worms from the same developmental stage. In addition, RNA was extracted from glp-4(bn2ts) mutant young adult worms. These glp-4 mutant adults contain somatic cells that form a sheath surrounding the gonad, but their germlines are developmentally restricted to just 12 cells stalled in prophase of mitosis when grown at restrictive temperature (Beanan and Strome 1992). This strain has been used extensively as a tool to identify changes in gene expression between germline and somatic tissues (Roussell and Bennett 1993; Shim 1999; Aoki et al. 2000; Higashitani et al. 2000; TeKippe and Aballay 2010). RT-PCR revealed a dramatic shift in top-1 pre-mRNA splicing from complete inclusion of the alternative exon in dissected gonads to partial inclusion in whole worms to almost complete skipping in dissected heads and in glp-4(bn2ts) worms grown at restrictive temperature (Fig. 1B). These data suggest top-1 cassette exon alternative splicing is strongly subject to tissue-specific regulation. This experiment also demonstrates a useful approach toward identifying germline-specific splicing events through comparison of alternative splicing between isolated gonad and whole glp-4(bn2ts) worms that lack an expanded germline.
Genome-wide identification of tissue-specific alternative splicing events
To identify tissue-specific alternative splicing events in the germline, we used high-throughput RNA sequencing and bioinformatics analysis of the resulting data. Total RNA was isolated from dissected wild-type gonads, as well as wild-type whole worms and glp-4(bn2ts) mutant whole worms 24 h past L4 stage (Fig. 2A). cDNA libraries were prepared and high-throughput sequencing was performed to obtain strand-specific, 50-bp paired-end reads. These reads were mapped to the C. elegans genome (ce6) with TopHat (Trapnell et al. 2009) and splicing changes were identified using SpliceTrap (Wu et al. 2011), which utilizes paired-end reads to quantify inclusion levels in alternative splicing events. We detected 23 events involving cassette exons with a minimum change in inclusion ratio (IR) of 0.3 between dissected gonad and glp-4 samples (Supplemental Table S1). The majority of alternative cassette exons we identified (19/23) are highly expressed in the glp-4 mutant, with some cassette exons nearly undetectable in gonad samples (Fig. 2B,C). Conversely, cassette exons from only a few genes (4/23) are included more often in RNA isolated from gonads when compared to glp-4 mutant RNA (Fig. 2D). Additionally, one gene contains an intron inclusion event enriched in RNA isolated from glp-4 adults that is only slightly detectable in wild-type dissected gonads.
Figure 2.
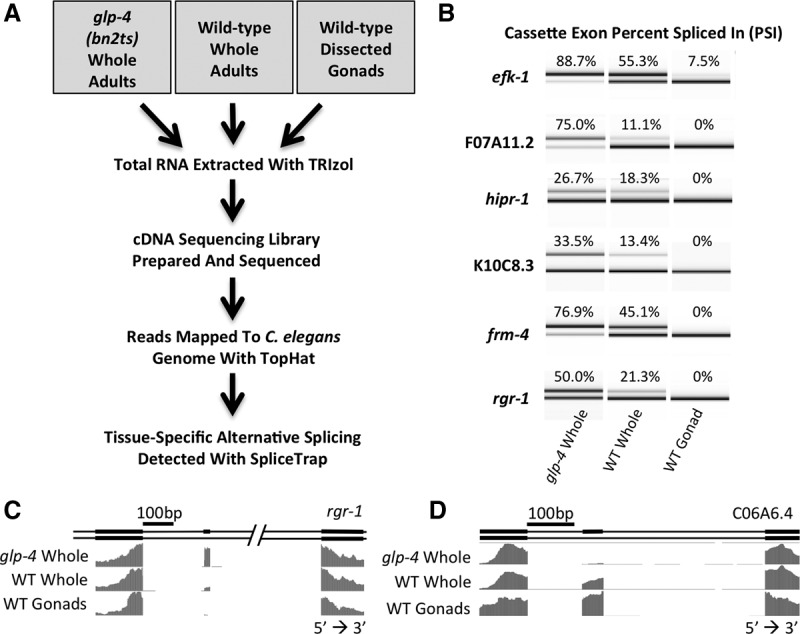
Cassette exon alternative splicing changes between C. elegans germline and somatic tissues. (A) Flowchart depicting the process of RNA isolation, high-throughput sequencing, and computational analysis. (B) RT-PCR products from primers that flank alternative cassette exons highlighting tissue-specific inclusion/skipping. Products were separated on an Agilent 2100 Bioanalyzer. PSI was calculated by dividing the molarity of the inclusion product by the sum of the molarities of the inclusion and skipping products. (C) Representation of the WormBase gene annotation for rgr-1 alternative isoforms and normalized RNA sequencing coverage tracks for the indicated libraries. This demonstrates skipping of the alternative cassette exon in gonad. (D) Representation of the WormBase gene annotation for C06A6.4 showing alternative isoforms and RNA sequencing coverage. This demonstrates an example of a cassette exon that is specifically included in the germline.
A shift from distal to proximal alternative 3′ splice site usage between the soma and the germline
Surprisingly, the majority of changes in alternative splicing we identified between germline-enriched and somatic-enriched samples were at adjacent alternative 3′ splice sites. Using SpliceTrap, we found 65 splice junctions in which adjacent 3′ splice sites (≤18 nt apart) are used in a tissue-specific manner (Supplemental Table S2). The threshold for tissue specificity was determined by identifying known adjacent 3′ splice sites with a minimum inclusion ratio difference of 0.3 between glp-4 and dissected wild-type gonads and a minimum of 15 junction-spanning reads in both libraries. In all of these 65 cases of adjacent alternative 3′ splice sites with tissue-specific usage, we observed a striking correlation: alternative splicing shifts from the distal 3′ splice site (furthest from the 5′ splice site) in the somatic (glp-4 whole worm) library toward a proximal 3′ splice site (closer to the 5′ splice site) in the germline library (Fig. 3A). As expected, RNA derived from wild-type whole worms, which contain both germline and somatic tissues, reveals an intermediate splicing pattern that uses a combination of proximal and distal 3′ splice sites. To verify the tissue-specific splicing patterns at 3′ splice sites seen in RNA-seq data, we isolated RNA from glp-4 adult whole worms, wild-type adult dissected heads, wild-type adult whole worms, and wild-type adult dissected gonads. We made cDNA via reverse transcription with random hexamers and then PCR-amplified with 32P-radiolabeled gene-specific primers that bind to sequences in exons flanking the alternative 3′ splice sites (Fig. 3B). The products were run on denaturing polyacrylamide gels and visualized with a PhosphorImager to verify the tissue-specific splicing at adjacent 3′ splice sites. Consistent with the RNA-seq data, glp-4 adult whole worm and wild-type adult dissected head samples showed a strong preference for the distal 3′ splice site. Wild-type adult whole worms produced isoforms that use a combination of proximal and distal 3′ splice sites, and wild-type adult germlines overwhelmingly produce isoforms that utilize the proximal 3′ splice site. Figure 3C shows the sequences of 15 representative adjacent alternative 3′ splice sites that show a shift to usage of the proximal site in the germline. Unlike NAGNAGs seen in mammals, in which the two adjacent 3′ splice sites are separated by 3 nt, the adjacent 3′ splice sites in C. elegans are separated by a short stretch of nucleotides enriched for pyrimidines (NAGYYYNAGs) (Fig. 3C). This is not surprising, given that nematodes differ from other eukaryotes in that they have a short intronic polypyrimidine tract immediately adjacent to the 3′ splice site with the consensus sequence YYYNAG (Aroian et al. 1993; Zhang and Blumenthal 1996). This requirement for immediately adjacent pyrimidines may preclude the ability of NAGNAG adjacent splice sites that are common in mammals to also function in worms.
Figure 3.
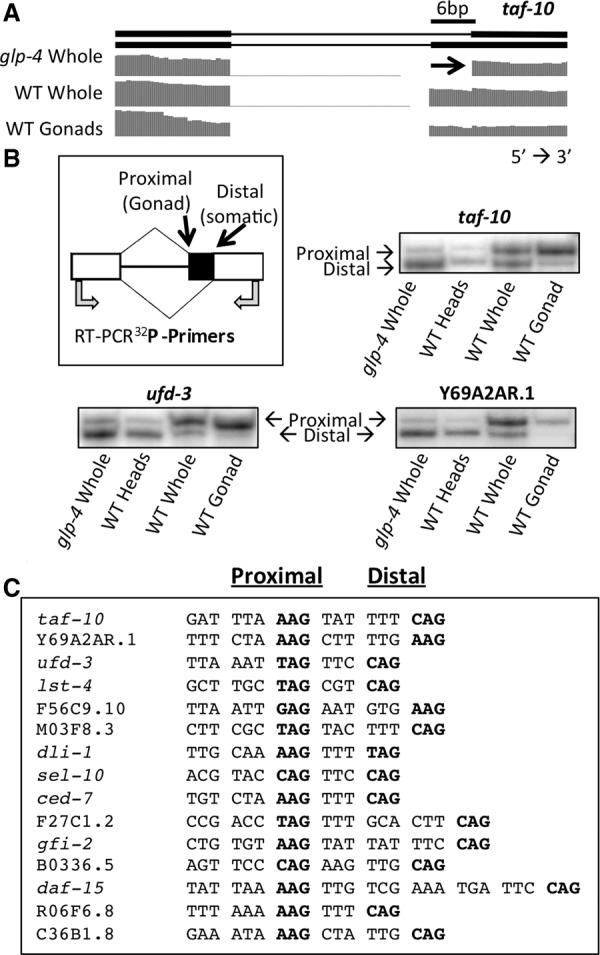
The major class of splicing changes between germline and somatic cells consists of adjacent alternative 3′ splice sites. (A) taf-10 alternative isoforms and RNA sequencing coverage tracks showing a tissue-specific alternative 3′ splicing event. Germline reads primarily cross the 3′ splice site closer to the 5′ splice site (proximal) while reads in somatic cells primarily use the 3′ splice site further from the 5′ splice site (distal). (B) Cartoon depicting the proximal and distal 3′ splice sites, the tissue-type in which each 3′ splice site is primarily used (in parentheses), and the location of the 32P-labeled oligos used to validate the tissue-specific enrichment of each isoform by RT-PCR. 32P RT-PCR products from three sample genes were separated on a 6% polyacrylamide denaturing gel and visualized with a PhosphorImager. (C) Nucleotides preceding and within tissue-specific 3′ splice sites in a representative set of genes. The proximal and distal 3′ splice sites are in bold for each sequence (left and right, respectively). Nucleotides are spaced to show the 3-nt periodicity and maintenance of frame between the 3′ splice sites.
Identification of additional introns with tissue-specific 3′ splice sites in the germline
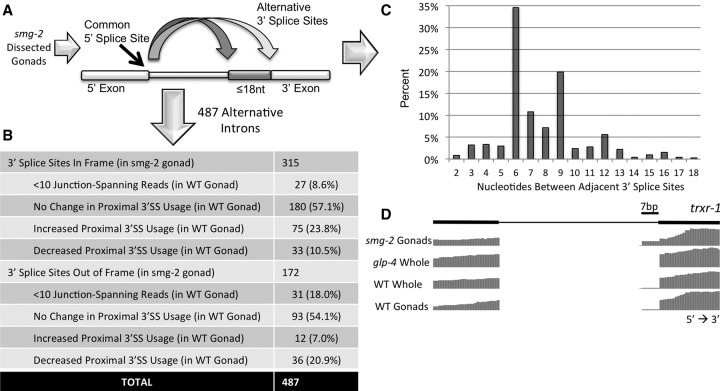
Because current genome annotations of alternative splicing are constantly evolving toward completion, we assumed that our identification of tissue-specific alternative 3′ splice sites using the SpliceTrap program, which depends on pre-annotation of alternative splicing, would be incomplete. Analysis that does not rely upon pre-annotation would allow us to identify novel tissue-specific splicing events from our sequencing data. When we measure alternative splicing by high-throughput sequencing or RT-PCR, we are measuring the steady-state levels of the alternative isoforms, which are influenced both by the splicing machinery and the relative stability of the different isoforms. Alternative isoforms with differing stabilities are seen in the phenomenon of alternative splicing coupled to nonsense-mediated decay (AS-NMD) (Hamid and Makeyev 2014). NMD is triggered when a message contains a premature termination codon (PTC), and in AS-NMD, a change in reading frame at the alternative splice junction generates a PTC. In C. elegans, unlike in other animals, worms bearing loss-of-function mutations in genes essential for the NMD pathway are viable (Hodgkin et al. 1989; Pulak and Anderson 1993), and this system offers us an opportunity to look for alternative adjacent 3′ splice sites that are not a multiple of 3 nt apart and would otherwise be destabilized by NMD because they disrupt reading frame. To understand the extent of adjacent tissue-specific 3′ splice sites genome-wide, and to understand if the NMD pathway is specifically involved in regulation of out-of-frame isoforms produced by adjacent 3′ splice sites, we dissected gonads from adult smg-2(e2008) worms defective in NMD (smg-2 is the C. elegans homolog of the essential NMD factor UPF1). We extracted total RNA and prepared cDNA libraries for high-throughput sequencing. We reasoned that smg-2 germline RNA would be enriched in NMD-target RNAs produced by alternative splicing (these would be preferentially degraded in wild-type germ cells), and by analyzing this sample we would maximize our chances of discovery of the different mRNA products produced by the spliceosome. RNA-seq reads were mapped to the C. elegans genome with TopHat (Trapnell et al. 2009) and introns with a common 5′ splice site but divergent 3′ splice sites ≤18 nt apart were identified (Fig. 4A).
Figure 4.
The majority of adjacent alternative 3′ splice sites are maintained in frame in the absence of nonsense-mediated decay (NMD). (A) Approach for identification of alternative 3′ splice site isoforms in the absence of NMD. Total RNA from smg-2 mutant gonads was extracted, made into a cDNA library, and subjected to high-throughput sequencing. Introns with common 5′ splice sites and adjacent alternative 3′ splice sites ≤18 nt apart were identified. (B) Table showing the number of alternative 3′ splice sites ≤18 nt apart in the smg-2 germline that maintain reading frame and those that cause a frameshift. This is further subdivided to show the corresponding change in expression or proximal splice site usage in the wild type relative to the smg-2 germline. (C) Graph depicting the percentage of introns (y-axis) identified in the smg-2 germline that have specified numbers of nucleotides separating the adjacent 3′ splice sites (x-axis). (D) Representation of the WormBase gene annotation for trxr-1. Note that alternative splicing is not annotated for this intron. The sequencing tracks show a frameshift-causing alternative 3′ ss 7 nt upstream with germline-specific isoform enrichment that is enhanced in the smg-2 mutant germline.
In analyzing smg-2 mutant RNA-seq data, we uncovered 487 unique introns with evidence of alternative 3′ splice sites ≤18 nt from each other (Supplemental Table S3). Of these alternative introns, 315 (64.7%) produced multiple isoforms in the same translational frame, while 172 (35.3%) resulted in alternative isoforms with a PTC-causing frameshift (Fig. 4B). This is consistent with previous studies that estimated that roughly 1/3 of alternative splicing events in C. elegans produce a PTC-containing isoform (Barberan-Soler et al. 2009). Cross-referencing these adjacent 3′ splice site introns detected in this smg-2 mutant germline analysis with splicing patterns in wild-type germline and glp-4 mutant whole worms, we identified 58 out of the 65 tissue-specific adjacent 3′ splice sites previously identified in the SpliceTrap analysis. In addition, we found 118 more introns that exhibit tissue specificity at adjacent 3′ splice sites using the same expression and splicing change thresholds as the previous analysis (Supplemental Table S2). Fifty three of 118 of these had no previously annotated alternative splicing, and these represent novel isoforms. We expect this to be an underestimation of alternative adjacent 3′ splice sites due to the requirement in our analysis that at least one read be detectable for each isoform in the smg-2 germline RNA-seq library; this requirement may have excluded tissue-specific alternative 3′ splice sites in which all reads that map to an intron in this library cross only the proximal splice site.
To ensure that it is, in fact, the presence of a mature germline that is leading to the production of proximal isoforms and not the mere lack of NMD, RNA-seq data from NMD-defective L1 (somatic tissue-enriched) larval worms from Kuroyanagi et al. (2013) were analyzed and compared to our NMD-deficient gonad sample data (Kuroyanagi et al. 2013). L1 smg-2 mutant splicing patterns for all 118 introns identified in our smg-2 gonad analysis mirrored that of glp-4 whole worms with the distal AG dinucleotide as the dominant 3′ splice site. Furthermore, a reciprocal analysis of common 5′ splice sites with alternative 3′ splice sites ≤18 nt apart in the smg-2 mutant L1 worm library failed to identify a single intron with adjacent 3′ splice sites that differed significantly in splicing pattern from glp-4. This suggests that it is an innate characteristic of the tissue, and not simply the lack of NMD, that contributes to the production and/or stability of the proximal isoforms in the smg-2 mutant animals.
Nonsense-mediated decay regulation of out-of-frame isoforms produced by adjacent 3′ splice sites
Many of the tissue-specific, out-of-frame proximal 3′ splice site isoforms expressed in the wild-type germline are found in the terminal exon (Supplemental Table S2), a situation not predicted to elicit an NMD response (Nagy and Maquat 1998). To determine if NMD regulation plays an extensive role in the degradation of germline-enriched proximal isoforms, we compared the relative expression and proximal 3′ splice site usage of all 487 smg-2 adjacent 3′ splice site introns to wild-type germline sequencing data (Fig. 4B). We note that a higher percentage of introns with out-of-frame proximal 3′ splice sites either drop below overall expression thresholds (minimum 10 junction-spanning reads) or decrease usage of the proximal 3′ splice site (minimum 0.3 PSI change) when compared to introns with in-frame proximal 3′ splice sites, suggesting that they are regulated by NMD. Figure 4D shows an example of this phenomenon with alternative adjacent 3′ splice sites separated by 7 nt in an intron of the trxr-1 gene. We conclude that even in the absence of NMD, the majority of alternative adjacent 3′ splice sites (54.0%) are found to be 6 nt or 9 nt from each other (Fig. 4C). Interestingly, out-of-frame adjacent 3′ splice sites are most commonly 7 nt or 8 nt apart, indicating that alternative adjacent 3′ splice sites are separated within a preferred range of 6–9 nt.
Proximal alternative 3′ splice site usage is favored in the germline
To better understand the characteristics of introns capable of tissue-specific alternative adjacent 3′ splice site usage in C. elegans, we further explored introns with an AG dinucleotide 6 nt upstream of a known 3′ splice site (NAGYYYNAGs). We used the UCSC Genome Browser Table Browser tool (Karolchik et al. 2004) to identify 106,891 annotated introns that terminate in an AG dinucleotide. From that set we then identified 1880 introns containing an AG dinucleotide 6 nt upstream of the terminal AG. We looked by hand at 1245 (66.2%) of these introns (Supplemental Table S4) to determine if these represent a rich source of adjacent alternative 3′ splice sites. By requiring a minimum expression threshold of 10 junction-spanning reads for an intron in both the wild-type germline and glp-4 whole worm libraries, we identified 192 introns for further study (Fig. 5A). Tissue-specific variation of proximal AG usage is evident when we arranged the 192 introns with sufficient expression in this study (>10 junction-spanning reads) according to increasing percentage of proximal 3′ splice site usage in the wild-type germline from 0% to 100% (Fig. 5B, black line) and then identified the corresponding percentage of proximal 3′ splice site usage for each intron in the somatic glp-4 library (Fig. 5B, gray line). Strikingly, 120 (62.5%) of these introns showed >10% usage of the upstream AG dinucleotide in either the soma or the germline. This indicates that the presence of an AG dinucleotide 6 nt upstream of the distal 3′ splice site, in a gene that is well expressed in the germline, is a strong indicator of potential adjacent alternative 3′ splice site usage. Conversely, of the 192 introns with sufficient expression, 72 used the proximal 3′ splice site minimally or not at all (<10% of junction-spanning reads) in glp-4, as expected, but also in the wild-type germline. This is despite the presence of an AG dinucleotide 6 nt upstream of the distal 3′ splice site.
Figure 5.
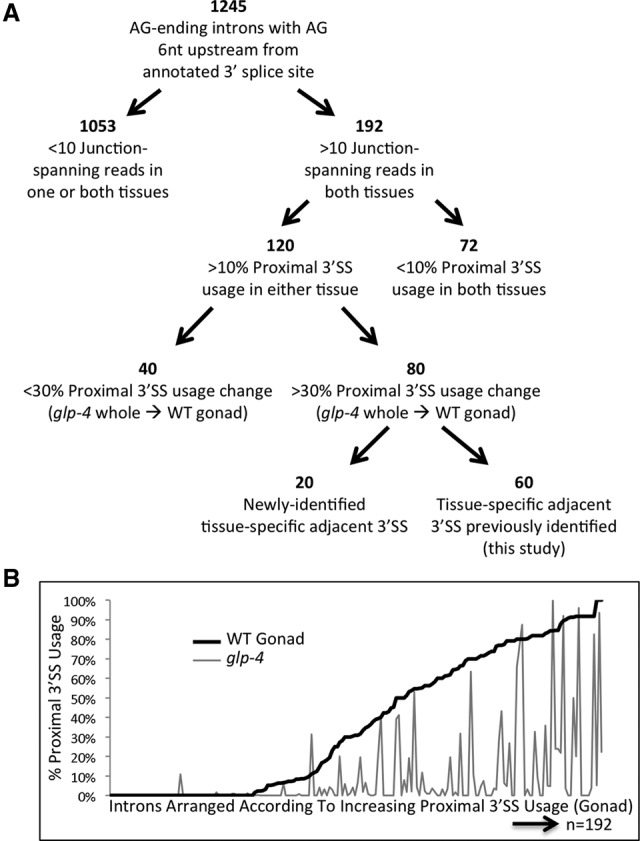
Proximal alternative 3′ splice site usage is favored in the germline. (A) Breakdown of annotated introns with a terminal AG dinucleotide and an AG dinucleotide 6 nt upstream. Introns separated according to the total number of reads crossing the splice junction, the presence of splicing at the proximal 3′ splice site, the change in percentage of isoforms using the proximal 3′ ss between glp-4 whole worm and wild-type germline, and the method used to identify the tissue-specific 3′ ss. (B) Graph of alternative adjacent proximal 3′ ss usage for the 192 introns with an AG dinucleotide 6 nt upstream of an annotated 3′ splice site and >10 junction-spanning reads in both germline and somatic libraries. Introns are arranged in order of increasing proximal 3′ splice site use percentage in the wild-type germline (black line, left to right) and the corresponding proximal 3′ splice site usage of each intron in the somatic glp-4 whole worm library (gray line).
Of the 120/192 with >10% proximal 3′ splice site usage, 80 showed tissue specificity (minimum 0.3 PSI change), with proximal isoform expression in the wild-type germline and distal isoform expression in glp-4. Sixty of these were previously identified in this study through SpliceTrap or smg-2 germline analysis, but 20 were novel discoveries of tissue-specific alternative 3′ splice sites. Using three methods (SpliceTrap, smg-2 gonad analysis, and this 6-nt Shift analysis), we have identified 203 alternative adjacent 3′ splice sites in total that show tissue-specific alternative splicing (Supplemental Table S2), all with a shift to the proximal splice site in the germline. It is important to note that for only a few cases did we detect proximal 3′ splice site usage in glp-4 roughly equivalent to the level of usage we observed in the wild-type germline. In fact, we have yet to identify a single intron in which there is a significant splicing change in the opposite direction (30% higher proximal splice site usage in glp-4 whole worm samples than in the wild-type germline).
Distance between the 5′ and 3′ splice sites correlates with the ability to detect splicing to the proximal 3′ splice site in the germline
In order to better understand why some introns with an AG dinucleotide 6 nt upstream of the annotated 3′ splice site do not splice to the proximal site in the germline while others do, we analyzed the lengths of introns in these two classes. C. elegans introns tend to be smaller on average than mammalian introns, though the factors that comprise the spliceosome tend to be well conserved. We compared the median length of introns that contain an AG dinucleotide 6 nt upstream that are either used or not used as a 3′ splice site in the germline (as previously defined in Fig. 5A). The class of introns that do not allow for proximal 3′ splice site usage in the germline have a median length of 49 nt, compared to a median length of 95 nt for introns with proximal 3′ splice site usage and a median length of 67 nt for 108,604 C. elegans introns identified in the UCSC Genome Browser (Fig. 6). This suggests that intron length may have influence over 3′ splice site choice, particularly in the context of adjacent 3′ splice sites. This is consistent with a model in which introns below a threshold length may not allow for splicing to a 6-nt proximal AG dinucleotide, even when expressed in the germline.
Figure 6.
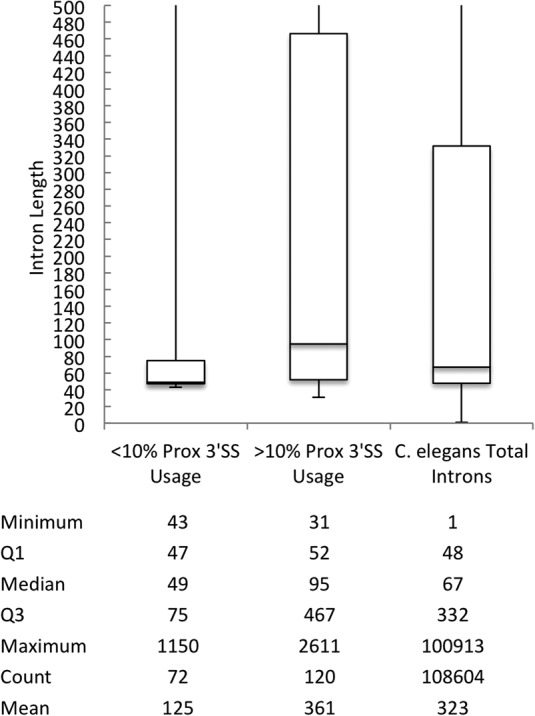
Introns that utilize the proximal AG dinucleotide as a 3′ splice site in the germline are longer than those that do not. Length of introns that utilize the proximal AG in <10% and >10% of junction-spanning reads as well as total C. elegans introns identified in the UCSC Genome Browser. The bottom and top of the boxes are the beginning of the second and fourth quartiles, respectively, with the median represented by the line in the center of the boxes.
Nucleotide content requirements for 3′ splice site selection differ between germline and somatic tissues
A large number of introns with an AG dinucleotide 6 nt upstream of the splice acceptor show adjacent alternative 3′ splice site usage in a tissue-specific manner. To understand the requirement for specific nucleotides in identifying a 3′ splice site in the context of these tissue-specific alternative adjacent 3′ splicing events, multiple sequence alignments of various intron classes (Supplemental Table S5) from the 3′ splice site to 40 nt upstream were analyzed through the online WebLogo program (Crooks et al. 2004). A random set of typical introns revealed the previously identified C. elegans 3′ splice site consensus motif TTTCAG (Zhang and Blumenthal 1996), while the consensus motif at tissue-specific alternative adjacent 3′ splice sites separated by 6 nt added only an AG dinucleotide consensus immediately upstream of the typical splice site motif (Fig. 7A,B, respectively). Surprisingly, the short stretch of pyrimidines that accompany the AG dinucleotide in the typical 3′ splice site motif are absent in the region upstream of the proximal AG dinucleotide preferred in the germline. Similarly, in consensus sequences from introns with 9 nt or 12 nt between tissue-specific alternative adjacent 3′ splice sites, the proximal AG dinucleotide is pushed 3 nt and 6 nt further upstream, respectively, with no further accompanying sequence consensus (Fig. 7C,D). Consensus sequence alignment motifs of introns in which the AG dinucleotide at the proximal 3′ splice site is minimally or not at all used in wild-type germline (Fig. 7E) do not reveal any major differences in nucleotide content when compared to introns in which usage of the proximal AG is enriched in the wild-type germline (Fig. 7B). This suggests that the proximal AG dinucleotide is not sufficient for germline 3′ splice site selection at adjacent 3′ splice sites and supports the influence of other factors such as intron length (Fig. 6). On the other hand, a consensus sequence alignment motif derived from introns with significant usage of the proximal AG in glp-4 whole worms (non-tissue-specific adjacent 3′ splice sites) shows the decreased presence of pyrimidines preceding the distal 3′ splice site AG dinucleotides with a concurrent increase preceding the proximal AG dinucleotide (Fig. 7F). Somatic cell-derived tissues may generally depend on nucleotide composition leading to U2AF binding upstream for alternative adjacent 3′ splice site decisions, while germline-specific use of upstream alternative 3′ splice sites correlates with a poor consensus sequence for the proximal site and a strong consensus for the distal site.
Figure 7.
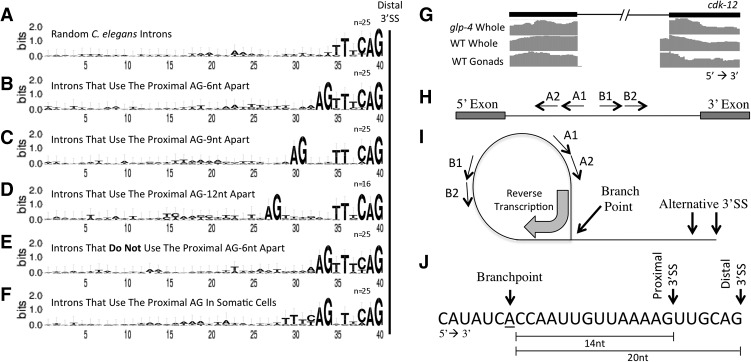
Sequence comparison and branchpoint analysis of introns with tissue-specific adjacent alternative 3′ splice sites. (A) WebLogo sequence comparison of regions upstream of a randomly selected set of 3′ splice sites. Upstream frequencies were measured from the distal splice site to 40 nt upstream. A logo displays the frequencies of bases at each position as the relative heights of letters, along with the degree of sequence conservation as the total height of a stack of letters, measured in bits of information. (B–D) WebLogo sequence comparisons of regions upstream of tissue-specific alternative 3′ splice sites that are 6 nt, 9 nt, and 12 nt apart. (E) WebLogo sequence comparison of tissue-specific alternative 3′ splice sites 6 nt apart in which the proximal AG dinucleotide is used <10% in the germline. Note the similarity of this sequence logo to that in which the dominant 3′ splice site in the germline is the proximal AG dinucleotide (Fig. 7B). (F) WebLogo sequence comparison of the 25 most used AG dinucleotide proximal 3′ splice sites in glp-4(bn2ts) when found 6 nt upstream of an intron-terminating AG dinucleotide. Note the signature of TTTCAG at the proximal 3′ splice site that is lacking in the tissue-specific proximal 3′ splice site (Fig. 7B). (G) Tissue-specific alternative 3′ splice site usage in the cdk-12 gene. (H) Orientation of divergent primers used to map branchpoints within an intron. (I) Lariat structure with alternative 3′ splice sites, the convergent orientation of the primers within the structure, and the branchpoint through which reverse transcriptase passes. Reverse transcription was first performed using the A1 primer, amplified with A1/B1 primers, purified using a Qiagen PCR clean-up kit, amplified a second time using the A2/B2 primers, and gel-purified. (J) Terminal 27 nt of the cdk-12 intron containing the tissue-specific alternative 3′ splice site with the proximal and distal 3′ splice sites and the location of the only branchpoint identified in our analysis. Also shown are the distances from this branchpoint to the proximal and distal 3′ splice sites.
An intron with a single branchpoint used in both soma and germline exhibits tissue-specific alternative adjacent 3′ splice site usage
One hypothesis to explain tissue-specific alternative 3′ splice site choice is that distinct intronic branchpoints are used in the tissues. Expansion or contraction of the distance between the branchpoint and NAGNAG 3′ splice sites in transgenes expressed in HEK293T cells was previously reported to enrich splice site selection at the proximal or distal 3′ splice site, respectively (Bradley et al. 2012). Conserved sequence motifs that typically mark intronic branchpoint locations in other eukaryotes have not been identified in C. elegans, making their identification more elusive. To assess whether a single branchpoint may be sufficient to allow for splicing at multiple adjacent 3′ splice sites, we set out to map the branchpoint(s) from an intron of cdk-12, which contains strongly tissue-regulated adjacent 3′ splice sites (Fig. 7G). RNA was extracted from glp-4 adults and wild-type gonads as previously described. A single, gene-specific primer complementary to the intron was used to reverse-transcribe through the branchpoint. cDNA from this reaction was amplified using divergent, nested primers within the intron (Fig. 7H). The PCR products were then ligated into plasmids, and 10–20 plasmid inserts were sequenced for each tissue type. A single branchpoint, 14 nt from the proximal 3′ splice site and 20 nt from the distal 3′ splice site, was observed for this intron of cdk-12 for both tissues examined (Fig. 7I). While an exhaustive, global mapping of C. elegans branchpoints has yet to be performed, these data provide evidence that the determination of the tissue-specific 3′ splice site for this intron occurs independently of the step in splicing at which the branchpoint is determined.
Adjacent alternative 3′ splice sites are conserved in related Caenorhabditis species
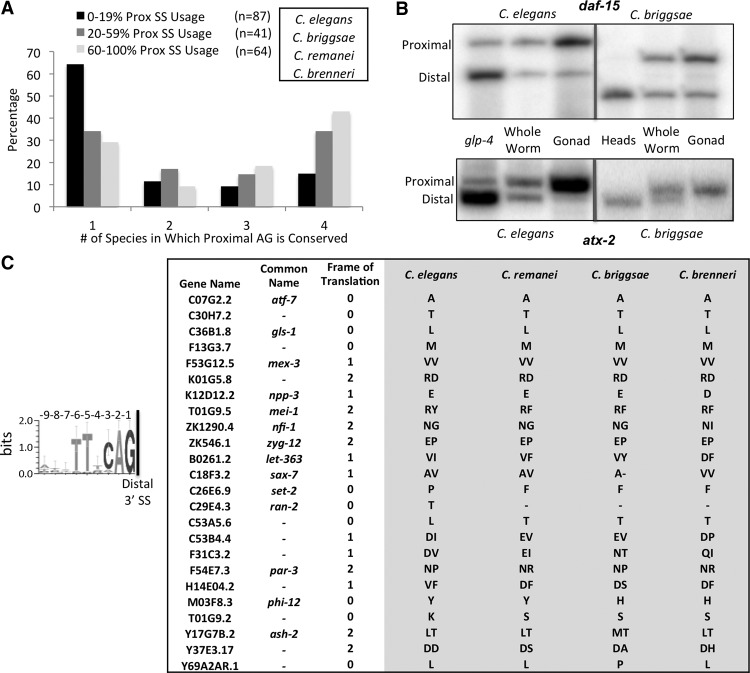
To test the evolutionary importance of alternative proximal 3′ splice site usage in the germline, we tested whether this tissue-specific phenomenon is conserved in related nematodes. The proximal AG dinucleotide is the only identifiable common sequence element in proximal 3′ splice sites in the C. elegans germline. Retention of this dinucleotide at these adjacent 3′ splice sites over evolutionary time in related nematode species C. briggsae, C. remanei, and C. brenneri would be indicative of conserved alternative splicing in these species. If alternative splicing is not conserved between the species, we would expect the proximal AG dinucleotide intronic sequence to change over evolutionary time, as intron sequences have been observed to change rapidly between Caenorhabditis species, with the exception of splicing regulatory regions (Kabat et al. 2006). Because evolutionary conservation typically suggests functional relevance, we compared the conservation of the proximal AG dinucleotide in species related to C. elegans to the degree of proximal AG dinucleotide usage as a 3′ splice site in the germline (Fig. 8A). For this, we evaluated the 192 introns with AG dinucleotides spaced 6 nt apart (Fig. 5A) that are sufficiently expressed in both the wild-type germline and glp-4 whole worm libraries. The vast majority of introns with low (0%–19%) proximal 3′ splice site usage in the germline do not exhibit proximal AG dinucleotide conservation in species related to C. elegans; 64% of the proximal AG dinucleotides from this class are only found in C. elegans, while only 15% are conserved across all four Caenorhabditis species. In contrast, introns with intermediate (20%–59%) and high (60%–100%) usage of the proximal 3′ splice site in the germline contain proximal AG dinucleotides more frequently in multiple nematodes species (34% and 43% are conserved in all four Caenorhabditis species, respectively). This shows that tissue-specific use of the AG dinucleotide in the germline in C. elegans is correlated with its conservation. Tissue specificity of splicing at adjacent 3′ splice sites may exist in these related nematode species.
Figure 8.
Adjacent alternative 3′ splice sites are conserved in related Caenorhabditis species. (A) Bar graph depicting conservation of the proximal AG dinucleotide in tissue-specific alternative 3′ splice sites. Introns were separated according to percent of proximal 3′ splice site usage in the wild-type germline (0%–19% in black, 20%–59% in dark gray, and 60%–100% in light gray). The comparison is made with three species related to C. elegans: C. remanei, C. briggsae, and C. brenneri. Displayed is a bar graph depicting the percentage of events in each category in which the upstream AG dinucleotide is conserved in only C. elegans (1) or in C. elegans plus one, two, or three other related species (2, 3, and 4). (B) 32P-labeled RT-PCR products made from primers that flank introns in daf-15 and atx-2 with tissue-specific alternative adjacent 3′ splice sites. Enrichment in C. elegans or C. briggsae for somatic cells (glp-4 or dissected heads, respectively) and germline cells (dissected gonads) shows a shift in abundance from the distal site isoform to the proximal site isoform in the germline of both species. (C) WebLogo consensus sequence motif of the 9 nt preceding the distal 3′ splice site of the 53 introns with tissue-specific 3′ splice sites separated by 9 nt (from Supplemental Table S2). Note the lack of conserved nucleotides at positions −7 to −9 from the distal 3′ splice site. On the right, a table depicting the 24 introns with the AG dinucleotide at the proximal site 9 nt upstream of the distal splice site conserved in all four nematode species analyzed. Shown is the gene name, common name, the frame of translation, and the identity of the amino acid(s) derived from the −7 to −9 nt.
To understand if splicing patterns at tissue-specific alternative 3′ splice sites in C. elegans are conserved among related species, we extracted total RNA from C. briggsae dissected heads, whole worms, and dissected gonads as in previous experiments. C. briggsae is a species of nematode that diverged from C. elegans roughly 50–100 million years ago (Coghlan and Wolfe 2002). The two species have maintained similar genome sizes and structure, but evolutionary changes in the number of genes and the amount of repetitive sequence exist (Stein et al. 2003). Comparative genomics approaches have more recently identified intronic sequence elements associated with alternative cassette exon splicing events conserved between C. elegans and C. briggsae (Kabat et al. 2006). RT-PCR with radiolabeled primers that anneal to flanking exons in atx-2 and daf-15 revealed conservation of tissue specificity at adjacent 3′ splice sites (Fig. 8B). As in C. elegans, splice site selection at these C. briggsae adjacent 3′ splice sites is directed to the proximal 3′ splice site in the germline and to the distal 3′ splice site in the heads, suggesting these tissue-specific splicing patterns are evolutionarily conserved.
Conservation across species of the amino acids added by alternative splicing is an important test of protein functionality. However, this is difficult to determine in the case of many adjacent alternative 3′ splice sites because the 6 nt upstream of the distal splice site match the C. elegans 3′ splice site consensus motif TTTCAG. While these nucleotides will likely be translated into amino acids if included by splicing at the proximal splice site in the germline, they already have evolutionary constraints on them to promote splicing at the distal site in somatic tissues. It is, therefore, difficult to determine if the amino acids added to the protein through alternative splicing to a proximal 3′ splice site 6 nt upstream in the gonad is important for protein function. However, for alternative adjacent 3′ splice sites that are separated by 9 nt, only the 6 nt immediately upstream of the proximal site would have these sequence constraints. This lack of positional nucleotide conservation can be observed in the WebLogo alignment of 53 tissue-specific alternative 3′ splice sites separated by 9 nt detailed in Supplemental Table S2 (Fig. 8C). The 3 nt upstream of the consensus motif (−7 through −9) do not have this evolutionary constraint for splicing. Of the 53 introns with adjacent 3′ splice sites separated by 9 nt, 24 show conservation of the proximal AG in all four nematode species analyzed, suggesting that the splicing pattern to these 3′ splice sites may also be conserved. Furthermore, the majority of amino acids derived from the −7 to −9 nucleotides within these 24 are either fully conserved among all four nematode species, conserved between two or more species, or the charge/polarity of the residues is conserved (Fig. 8C). The conservation of these amino acids suggests that the use of tissue-specific alternative adjacent 3′ splice sites corresponds with conservation of changes in the primary structure of the alternative protein isoforms produced.
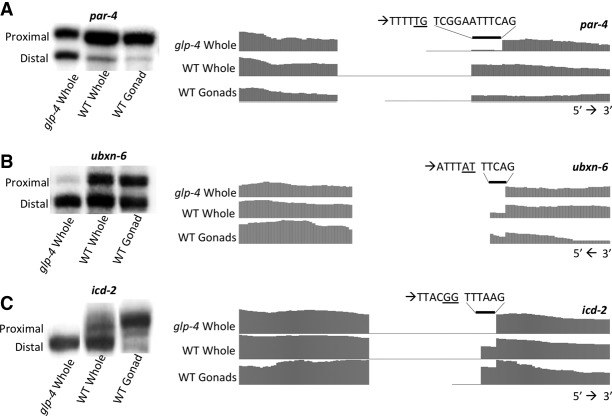
Noncanonical alternative adjacent 3′ splice acceptor dinucleotides are used in the wild-type germline
Previously, the TTTC polypyrimidine tract was found to be sufficient to induce splicing following mutation of a TTTCAG 3′ splice site to TTTCAA (Zhang and Blumenthal 1996), indicating that cryptic splicing could occur at non-AG dinucleotide acceptors. In our analyses of tissue-specific adjacent 3′ splice sites, we identified three introns in C. elegans in which the germline-preferred proximal 3′ splice site does not end in the canonical AG (Fig. 9A–C; Supplemental Table S2). Previous EST evidence demonstrates splicing to these sites, but this work provides the first evidence that this splicing is done tissue-specifically in germline cells and is not a result of cryptic splicing from mutation of a native AG acceptor. In these examples, splicing at TG, AT, and GG dinucleotide acceptors (par-4, ubxn-6, and icd-2, respectively) is observed and strongly preferred in the germline, despite the apposition of 3′ splice sites used heavily in somatic cells containing strong consensus sequence motifs. This small number of cases further delineates the deviation of 3′ splice site choice in germline cells from canonical 3′ splice site dinucleotide composition requirements found in somatic tissues.
Figure 9.
3′ splice sites in dissected gonad samples that deviate from the canonical AG dinucleotide. 32P RT-PCR and RNA sequencing coverage tracks of handpicked glp-4 whole worm, wild-type whole worm, and wild-type germline RNA showing tissue-specific expression of isoforms with alternative 3′ splice sites. Analysis of par-4 (A), ubxn-6 (B), and icd-2 (C) RNA sequencing reveals significant usage of noncanonical 3′ splice sites primarily in germline-containing samples. Sequences above each set of coverage tracks show the dinucleotide set found at the somatic site preceded by the dinucleotide set found at the germline site (underlined) and the accompanying sequence immediately upstream. Genomic and mRNA sequences in these regions were validated.
Discussion
Adjacent alternative 3′ splice sites have been identified in several organisms and tissues. Studies in humans and mice as well as plants have inspired debate concerning how these tandem splice sites are chosen. Statistical and predictive computational models trained on human data sets propose splicing between adjacent 3′ splice sites is randomly distributed or relies strictly on the content of nucleotides within the sites (Sinha et al. 2009). These models struggled with the unique structure of the 3′ splice site-determining cis-elements in C. elegans introns. It was also perceived that the lack of a clear branchpoint consensus sequence and long polypyrimidine tract in C. elegans would not allow for alternative splicing at tandem acceptor sites (Hiller et al. 2004).
Our work shows that adjacent alternative 3′ splice sites are present and functional in C. elegans as well as in its nematode relative C. briggsae. Furthermore, we show that splicing patterns to these sites are regulated and tissue-specific. In this study, we identify alternative splicing differences between C. elegans somatic and germline tissues. Through RNA sequencing of isolated gonads as well as glp-4(bn2ts) mutant worms that fail to produce a functional, expanded germline, we used pre-annotated and de novo transcriptomes to identify 23 cassette exons, one intron retention, and 203 adjacent 3′ splice sites that are differentially spliced between the two tissue types. A large portion of these tissue-regulated alternative 3′ splice sites differ by 6 or 9 nt, even in an NMD abrogated background. There is evidence that this preservation of frame is not by chance, as mutations that favor frame preservation at Drosophila NAGNAGs allows splicing regulation divergence to be more easily tolerated (McManus et al. 2014).
The distal splice site used in somatic tissues mirrors a consensus 3′ splice site in C. elegans with an AG dinucleotide preceded by a short stretch of pyrimidines. The frequency of splicing to the proximal site, which is normally preferred in germline samples, increases in somatic samples when a proximal AG dinucleotide is preceded by a stretch of pyrimidines (i.e., when the proximal site is composed of nucleotides that make up a traditional 3′ splice site). This indicates that 3′ splice site selection in somatic tissues relies on the TTTCAG 3′ splice site consensus. In an effort to identify if there are sequence clues that drive splicing to the proximal site in germline cells, we compared the frequency of 4-nt, 5-nt, and 6-nt DNA words in introns containing tissue-specific alternative adjacent 3′ splicing against total introns (data not shown). We did this with either the 50 nt closest to the 3′ end of the introns (the region upstream of the proximal AG) or with total introns. We could find no evidence of consensus sequence motifs that were enriched in introns undergoing tissue-specific alternative adjacent 3′ splicing. This differed markedly from our previous analysis of intron sequences flanking alternative cassette exons, where words enhanced in these introns relative to total introns matched known components of the splicing code and allowed for identification of new ones (Kabat et al. 2006).
Introns in C. elegans tend to be relatively small with a median length of 67 nt (Fig. 6). At some minimal distance, spatial constraints of the spliceosome will likely dictate limits on the branchpoints and splice sites used in the excision of the intron. The contribution to 3′ splice site choice that the physical distance from the alternative 3′ splice site to the branchpoint makes has been explored in other organisms (Akerman and Mandel-Gutfreund 2006; Tsai et al. 2007, 2010; Bradley et al. 2012). Expression analysis of a PTB minigene with 4-nt or 7-nt insertions between the branchpoint and tandem acceptor set caused an increase in splicing to the proximal site, suggesting that the choice of branchpoint may influence the choice of 3′ splice site at NAGNAGs (Tsai et al. 2010). Conversely, tissue-specific branchpoint mapping in this study revealed no alternative branchpoint usage in an intron with adjacent tissue-specific 3′ splice sites (Fig. 7). Instead, introns with activated proximal AG dinucleotides in the germline tend to be significantly longer overall (Fig. 6) and possibly more permissive of spliceosomal spatial modulations that lead to 3′ splice site choice fluctuations. If alternative branchpoints do not lead to alternative 3′ splice sites, perhaps the scanning mechanism from branchpoint to splice acceptor differs between the germline and somatic splicing machineries.
The discovery of so many tissue-specific alternative adjacent 3′ splice sites, in which the proximity to the 5′ splice site is the main determinant of alternative splicing in the germline, suggests that there is an overall difference in 3′ splice site selection in the germline relative to other tissues. Differential expression of splicing trans-factors between germline and somatic tissues that are part of the U2 snRNP or are interactors with the U2AF subunit homologs UAF-1 and UAF-2 could explain this phenomenon. To begin to address this, we used a database of 494 spliceosome-associated components (Cvitkovic and Jurica 2013) to identify spliceosomal proteins and RNA binding proteins in C. elegans. Cross-referencing our tissue-specific RNA-seq data using the DESeq analysis software (Anders and Huber 2010), we identified candidate factors enriched or depleted in the germline relative to somatic cells (Supplemental Table S6; Supplemental Fig. 1). Two candidates to look at, UAF-1 and UAF-2, did not yield expression changes of significance in this analysis, and no detectable change in splicing pattern at adjacent 3′ splice sites was observed in a mutant allele (uaf-1) or following RNAi knockdown (uaf-2) (Supplemental Fig. 2). Similarly, we tested other candidate splicing regulators, including C. elegans mog-2 (mammalian U2A′ homolog), ptb-1 (SAP49; polypyrimidine tract-binding protein), hrpf-1 and sym-2 (hnRNP H/F homologs), and sfa-1 (branchpoint-binding protein homolog), and detected no change in splicing patterns at tissue-specific 3′ splice sites. One possibility is that splicing fidelity is lower in the germline and proofreading mechanisms may not be as robust. This could explain the use of proximal sites that lack the YYYNAG consensus and even proximal sites that lack the AG dinucleotide.
Our data indicate that the presence of an AG dinucleotide 6–9 nt upstream of a strong consensus 3′ splice site will likely lead to proximal alternative splice site usage if the pre-mRNA is expressed in the germline and the intron is above a minimal length. Importantly, our data indicate that these tissue-specific alternative splicing events, their component nucleotides, and the resulting amino acids within the exonic extensions are conserved in related nematode species. No matter the mechanism by which germline-specific splicing is established at adjacent 3′ splice sites, this mechanism has been adapted to drive conserved alternative splicing events.
Methods
Strain maintenance and dissections
Strains: glp-4(bn2ts), smg-2(e2008), and wild-type (Bristol N2 strain) worms were obtained from the Caenorhabditis Genetics Center. Synchronized worm populations were obtained by axenization of a mixed population of worms cultured on liquid media at 20°C to isolate embryos (Lewis and Fleming 1995). After synchronization, all worms were cultured at 25°C, the restrictive temperature for glp-4(bn2ts), and the developmental stage was confirmed by microscopy of vulva development (white-crescent stage).
RNA extraction for RT-PCRs
Gonads were dissected on slides in 50 µL dissection buffer (110 µL 10× Egg Buffer [1 M HEPES, 5 M NaCl, 1 M MgCl2, 1 M CaCl2, 1 M KCl], 25 µL 20% TWEEN, 100 µL 10 mM levamisole, 865 µL ddH2O) with 30-gauge needles. One hundred to 150 heads were removed by cutting below the pharynx. Seventy-five to 100 extruded gonads were isolated by removing the heads and cutting at the spermatheca. One hundred to 200 adult worms 20–24 h past L4 were used for whole worm samples. Tissues were placed into 300 µL TRIzol (Invitrogen) on ice. Sixty microliters chloroform were added to the TRIzol/tissue samples, and phases were separated using pre-spun Phase Lock Gel-Heavy tubes (5 Prime). The aqueous phase was transferred to a new tube and mixed with 125–175 µL isopropanol and 1 µL GlycoBlue (Ambion). RNA was precipitated overnight at −20°C and pelleted at 13,000g at 4°C for 30 min. The pellet was washed with 75% ethanol, dried, and resuspended in 10–20 µL ddH2O.
Reverse transcription
One to 10 µL of RNA was mixed with 1 µL 10 mM dNTPs, 1 µL 50 µM random hexamers, and ddH2O to 13 µL. The solution was incubated at 65°C for 5 min and then for 1 min on ice. Four microliters first-strand buffer, 2 µL DTT, and 1 µL SuperScript III (Invitrogen) were added, and the following incubation protocol was followed: 4°C 10 min, 15°C 10 min, 42°C 20 min, 70°C 15 min.
Polymerase chain reaction
For cassette exons, gene-specific primers (10 µM) that anneal to sequences in exons that flank an alternative exon were used in a PCR reaction with 1–2 µL cDNA from RT reactions. The annealing temperature was 50°C and the elongation time was 30 sec with 25–28 cycles of amplification. Primer sequences are available in Supplemental Table S8. One microliter of product was run on an Agilent 2100 Bioanalyzer with a DNA 1000 kit and/or separated on a 2% agarose gel. Dividing the molarity of the inclusion isoform by the sum of molarities of inclusion and skipping isoforms determined the percent spliced in (PSI). For alternative 3′ splice sites, PCR reactions were run similar to cassette exons, but the elongation time was 15 sec and the reactions were run in the presence of 32P-labeled reverse primer. Radiolabeled PCR products from alternative 3′ splice sites were phenol, then chloroform extracted and ethanol precipitated at −20°C overnight. After microcentrifugation, the pellets were resuspended in 10 µL formamide dye, heated to 95°C, and run on a long 6% polyacrylamide urea denaturing gel at 1700V/45W for 3.5–4 h. This gel was exposed to a PhosphorImager screen overnight and bands were imaged on a Typhoon scanner.
RNA-seq
RNA extractions were performed with TRIzol (Invitrogen) and further purified using the RNeasy Plus Micro kit (Qiagen). Gonad samples were depleted of rRNA with a RiboZero kit (Epicentre), and whole worm samples were poly(A) selected. mRNA sequencing libraries were sequenced according to manufacturer recommendations with the TruSeq Stranded mRNA Sample kit (Illumina) at the Centre for Genomic Regulation sequencing facility in Barcelona, Spain. A total of 50-bp paired-end reads were mapped to the C. elegans reference genome (ce6) with TopHat (Trapnell et al. 2009), and PCR duplicates were removed. SpliceTrap (Wu et al. 2011) was used to detect pre-annotated splicing changes between tissues that met the threshold of a 0.3-minimum inclusion ratio change and a minimum count of 15 junction-spanning reads (either skipping or inclusion on each side). Identification of smg-2(e2008) alternative 3′ splice sites was performed on dissected gonad sample libraries. Splice junctions were extracted from TopHat mappings and code was written to identify introns that were filtered according to (1) introns with more than one 3′ splice site for the same 5′ splice site, and (2) among the 3′ splice sites for a given 5′ splice site, the distance between 3′ splice sites is ≤18 nt. The resulting introns were hand-curated to detect tissue-specific splicing changes at adjacent 3′ splice sites.
Consensus motifs
Typical nonalternatively spliced introns were selected by random gene and intron selection in the UCSC Genome Browser. Intron sequences contain the previously annotated 3′ splice site through 40 nt upstream. A consensus motif was created online with WebLogo (http://weblogo.threeplusone.com/create.cgi). Twenty-five introns from the following classes—(1) 6-nt, 9-nt, and 12-nt shifted alternative 3′ splice sites, (2) 3′ splice sites in which the proximal AG is unused in gonad samples, and (3) often used 3′ splice sites in glp-4(bn2ts)—were chosen, and consensus motifs were created in the same manner as typical 3′ splice sites.
Branchpoint identification
RNA was extracted from glp-4 whole worms or dissected wild-type gonads. Divergent nested primer sets (A1/B1 and A2/B2) were designed to anneal to an intron within cdk-12 that contains a tissue-specific alternative 3′ splice site. Reverse transcription was performed as previously stated substituting the A1 primer for random hexamers. cDNA from this reaction was amplified in a primary PCR reaction with primers A1 and B1 (28 cycles), purified in a PCR Clean-up kit (Qiagen), and reamplified in a secondary PCR reaction using primers A2 and B2 (25 cycles). Primer sequences are available in Supplemental Table S8. The resulting amplicon was gel purified, cloned into pCR2.1 TOPO (Invitrogen), and transformed into DH5alpha chemically competent cells. A minimum of 12 colonies were selected for sequencing each from the samples representing glp-4 adult RNA and wild-type gonad RNA.
Intron length analysis
Nucleotide lengths were measured for introns with >10 junction-spanning reads and <10% (n = 72) or >10% (n = 120) proximal 3′ ss usage in the germline (Fig. 5). In addition, introns were identified in the UCSC Genome Table Browser (ce6 release), and nucleotide lengths were measured. The minimum, first quartile, median, third quartile, and maximum lengths were determined from each set and compared.
Data access
FASTQ files of raw reads from five high-throughput RNA-seq libraries and a table of genes with tissue-specific alternative 3′ splice sites have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession number GSE64672.
Acknowledgments
We thank Juan Valcárcel and the Genomics Unit of the Centre for Genomic Regulation in Barcelona, who provided valuable discussions as well as support for cDNA preparation and high-throughput sequencing. We also thank our UC Santa Cruz colleagues Manny Ares, Melissa Jurica, Jeremy Sanford, Needhi Bhalla, and Susan Strome for advice and helpful discussions. Strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This research is supported by a grant from the National Science Foundation, MCB-1121290, to A.M.Z.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.186783.114.
References
- Akerman M, Mandel-Gutfreund Y. 2006. Alternative splicing regulation at tandem 3′ splice sites. Nucleic Acids Res 34: 23–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki H, Sato S, Takanami T, Ishihara T, Katsura I, Takahashi H, Higashitani A. 2000. Characterization of Ce-atl-1, an ATM-like gene from Caenorhabditis elegans. Mol Gen Genet 264: 119–126. [DOI] [PubMed] [Google Scholar]
- Ares M Jr, Weiser B. 1995. Rearrangement of snRNA structure during assembly and function of the spliceosome. Prog Nucleic Acid Res Mol Biol 50: 131–159. [DOI] [PubMed] [Google Scholar]
- Aroian RV, Levy AD, Koga M, Ohshima Y, Kramer JM, Sternberg PW. 1993. Splicing in Caenorhabditis elegans does not require an AG at the 3′ splice acceptor site. Mol Cell Biol 13: 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S, Ragle JM. 2013. Alternative splicing regulation of cancer-related pathways in Caenorhabditis elegans: an in vivo model system with a powerful reverse genetics toolbox. Int J Cell Biol 2013: 636050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S, Zahler AM. 2008. Alternative splicing regulation during C. elegans development: splicing factors as regulated targets. PLoS Genet 4: e1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S, Lambert NJ, Zahler AM. 2009. Global analysis of alternative splicing uncovers developmental regulation of nonsense-mediated decay in C. elegans. RNA 15: 1652–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S, Fontrodona L, Ribo A, Lamm AT, Iannone C, Ceron J, Lehner B, Valcarcel J. 2014. Co-option of the piRNA pathway for germline-specific alternative splicing of C. elegans TOR. Cell Rep 8: 1609–1616. [DOI] [PubMed] [Google Scholar]
- Beanan MJ, Strome S. 1992. Characterization of a germ-line proliferation mutation in C. elegans. Development 116: 755–766. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. 1997. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89: 781–787. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Abovich N, Rosbash M. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev 12: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T, Steward K. 1997. RNA processing and gene structure. In C. elegans II (ed. Riddle DL, et al. ), pp. 117–145. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Bradley RK, Merkin J, Lambert NJ, Burge CB. 2012. Alternative splicing of RNA triplets is often regulated and accelerates proteome evolution. PLoS Biol 10: e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Cha DS, Hollis SE, Datla US, Lee S, Ryu J, Jung HR, Kim E, Kim K, Lee M, Li C, et al. 2012. Differential subcellular localization of DNA topoisomerase-1 isoforms and their roles during Caenorhabditis elegans development. Gene Expr Patterns 12: 189–195. [DOI] [PubMed] [Google Scholar]
- Coghlan A, Wolfe KH. 2002. Fourfold faster rate of genome rearrangement in nematodes than in Drosophila. Genome Res 12: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvitkovic I, Jurica MS. 2013. Spliceosome database: a tool for tracking components of the spliceosome. Nucleic Acids Res 41: D132–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid FM, Makeyev EV. 2014. Emerging functions of alternative splicing coupled with nonsense-mediated decay. Biochem Soc Trans 42: 1168–1173. [DOI] [PubMed] [Google Scholar]
- Higashitani A, Aoki H, Mori A, Sasagawa Y, Takanami T, Takahashi H. 2000. Caenorhabditis elegans Chk2-like gene is essential for meiosis but dispensable for DNA repair. FEBS Lett 485: 35–39. [DOI] [PubMed] [Google Scholar]
- Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. 2004. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat Genet 36: 1255–1257. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollins C, Zorio DA, MacMorris M, Blumenthal T. 2005. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA 11: 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabat JL, Barberan-Soler S, McKenna P, Clawson H, Farrer T, Zahler AM. 2006. Intronic alternative splicing regulators identified by comparative genomics in nematodes. PLoS Comput Biol 2: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H, Kimura T, Wada K, Hisamoto N, Matsumoto K, Hagiwara M. 2000. SPK-1, a C. elegans SR protein kinase homologue, is essential for embryogenesis and required for germline development. Mech Dev 99: 51–64. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Watanabe Y, Suzuki Y, Hagiwara M. 2013. Position-dependent and neuron-specific splicing regulation by the CELF family RNA-binding protein UNC-75 in Caenorhabditis elegans. Nucleic Acids Res 41: 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Jang YJ, Koo HS. 1998. Alternative splicing in the Caenorhabditis elegans DNA topoisomerase I gene. Biochim Biophys Acta 1396: 207–214. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Fleming JT. 1995. Basic culture methods. Methods Cell Biol 48: 3–29. [PubMed] [Google Scholar]
- McManus CJ, Coolon JD, Eipper-Mains J, Wittkopp PJ, Graveley BR. 2014. Evolution of splicing regulatory networks in Drosophila. Genome Res 24: 786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Nagy E, Maquat LE. 1998. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 23: 198–199. [DOI] [PubMed] [Google Scholar]
- Pulak R, Anderson P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 7: 1885–1897. [DOI] [PubMed] [Google Scholar]
- Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, Lee LJ, Morris Q, Blencowe BJ, Zhen M, et al. 2011. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res 21: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussell DL, Bennett KL. 1993. glh-1, a germ-line putative RNA helicase from Caenorhabditis, has four zinc fingers. Proc Natl Acad Sci 90: 9300–9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim YH. 1999. elt-1, a gene encoding a Caenorhabditis elegans GATA transcription factor, is highly expressed in the germ lines with msp genes as the potential targets. Mol Cells 9: 535–541. [PubMed] [Google Scholar]
- Sinha R, Nikolajewa S, Szafranski K, Hiller M, Jahn N, Huse K, Platzer M, Backofen R. 2009. Accurate prediction of NAGNAG alternative splicing. Nucleic Acids Res 37: 3569–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Porro EB, Patton JG, Nadal-Ginard B. 1989. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature 342: 243–247. [DOI] [PubMed] [Google Scholar]
- Smith CW, Chu TT, Nadal-Ginard B. 1993. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol 13: 4939–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J, Lawson D. 2006. Overview of gene structure. In WormBook (ed. The C. elegans Research Community), pp. 1–10. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. 2003. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol 1: E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56: 110–156. [DOI] [PubMed] [Google Scholar]
- TeKippe M, Aballay A. 2010. C. elegans germline-deficient mutants respond to pathogen infection using shared and distinct mechanisms. PLoS One 5: e11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KW, Tarn WY, Lin WC. 2007. Wobble splicing reveals the role of the branch point sequence-to-NAGNAG region in 3′ tandem splice site selection. Mol Cell Biol 27: 5835–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai KW, Chan WC, Hsu CN, Lin WC. 2010. Sequence features involved in the mechanism of 3′ splice junction wobbling. BMC Mol Biol 11: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB. 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Akerman M, Sun S, McCombie WR, Krainer AR, Zhang MQ. 2011. SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics 27: 3010–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM. 2012. Pre-mRNA splicing and its regulation in Caenorhabditis elegans. In WormBook (ed. The C. elegans Research Community), pp. 1–21. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Blumenthal T. 1996. Functional analysis of an intron 3′ splice site in Caenorhabditis elegans. RNA 2: 380–388. [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. 1992. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci 89: 8769–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. 1999. Both subunits of U2AF recognize the 3′ splice site in C. elegans. Nature 402: 835–838. [DOI] [PubMed] [Google Scholar]