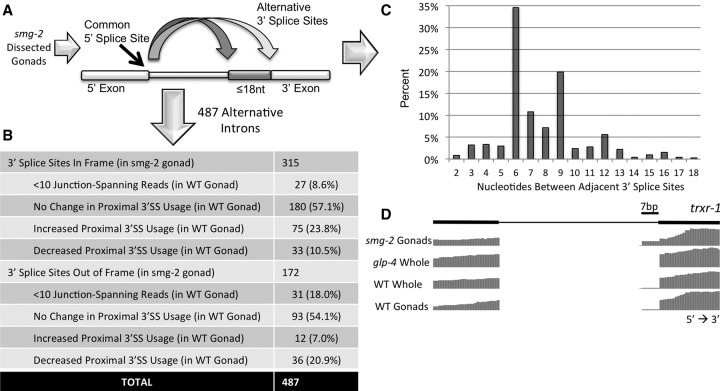
Figure 4.
The majority of adjacent alternative 3′ splice sites are maintained in frame in the absence of nonsense-mediated decay (NMD). (A) Approach for identification of alternative 3′ splice site isoforms in the absence of NMD. Total RNA from smg-2 mutant gonads was extracted, made into a cDNA library, and subjected to high-throughput sequencing. Introns with common 5′ splice sites and adjacent alternative 3′ splice sites ≤18 nt apart were identified. (B) Table showing the number of alternative 3′ splice sites ≤18 nt apart in the smg-2 germline that maintain reading frame and those that cause a frameshift. This is further subdivided to show the corresponding change in expression or proximal splice site usage in the wild type relative to the smg-2 germline. (C) Graph depicting the percentage of introns (y-axis) identified in the smg-2 germline that have specified numbers of nucleotides separating the adjacent 3′ splice sites (x-axis). (D) Representation of the WormBase gene annotation for trxr-1. Note that alternative splicing is not annotated for this intron. The sequencing tracks show a frameshift-causing alternative 3′ ss 7 nt upstream with germline-specific isoform enrichment that is enhanced in the smg-2 mutant germline.