Figure 5.
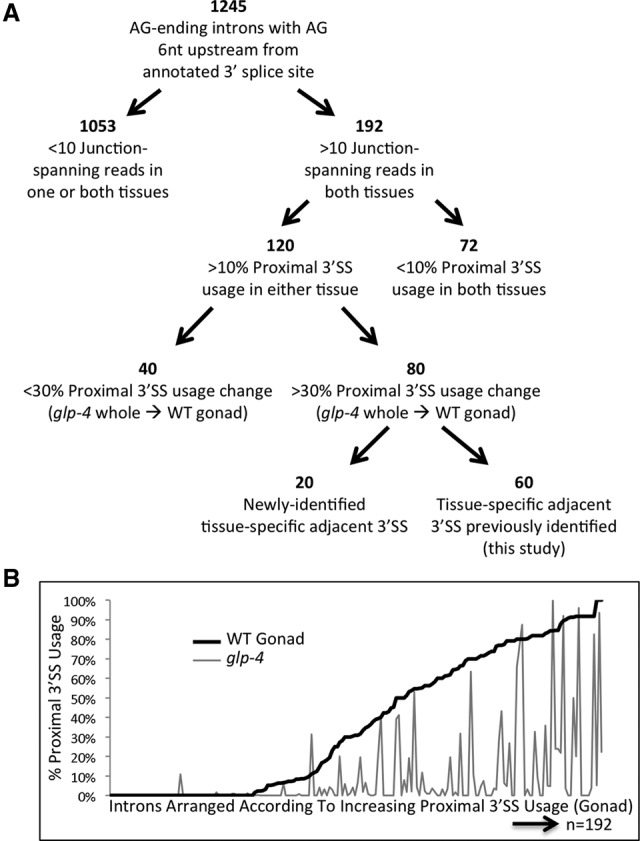
Proximal alternative 3′ splice site usage is favored in the germline. (A) Breakdown of annotated introns with a terminal AG dinucleotide and an AG dinucleotide 6 nt upstream. Introns separated according to the total number of reads crossing the splice junction, the presence of splicing at the proximal 3′ splice site, the change in percentage of isoforms using the proximal 3′ ss between glp-4 whole worm and wild-type germline, and the method used to identify the tissue-specific 3′ ss. (B) Graph of alternative adjacent proximal 3′ ss usage for the 192 introns with an AG dinucleotide 6 nt upstream of an annotated 3′ splice site and >10 junction-spanning reads in both germline and somatic libraries. Introns are arranged in order of increasing proximal 3′ splice site use percentage in the wild-type germline (black line, left to right) and the corresponding proximal 3′ splice site usage of each intron in the somatic glp-4 whole worm library (gray line).
