Figure 8. A reduction in Cx43 channel activity increases migration.
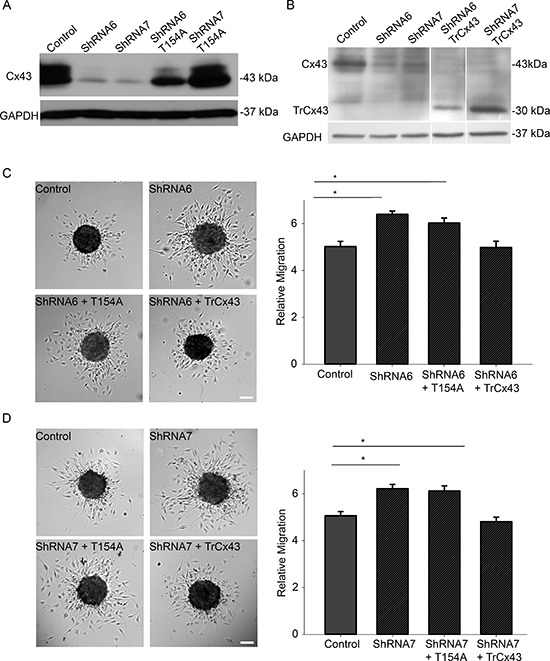
Western blot analysis was done to examine the successful expression of mutants T154A and TrCx43 in ShRNA6 and ShRNA7 cells. (A) We used anti-Cx43 (Sigma) that targets the C-terminal domain of Cx43 to detect mutant protein T154A. Using densitometry (ImageJ) we found Cx43 expression in control cells to be 16 times higher than ShRNA6 and ShRNA7 cells. Cx43 expression in ShRNA6-T154A and ShRNA7-T154A cells was 9 and 16 times higher, respectively. (B) Since the TrCx43 mutant lacks the C-terminal tail we could not use the Sigma anti-Cx43 antibody we used for detection of wild-type Cx43 or T154A. We used P1E11 anti-Cx43 antibody (Fred Hutchinson Cancer Research Center) that targets the N-terminal of Cx43 to detect TrCx43 expression in ShRNA6 and ShRNA7 cells. Since the TrCx43 is lacking the C-terminal tail it is of a smaller molecular weight, predicted to be 30 kDa. Cx43 expression in control cells was 6 times higher than ShRNA6 and ShRNA7 cells. Cx43 expression in ShRNA6-TrCx43 and ShRNA7-TrCx43 cells was 3 and 6 times higher, respectively. Please note that extraneous lanes were removed from the blot. The expression of T154A mutant in the ShRNA6 and shRNA7 cells did not reduce migration levels to that of control cells. An increase in migration comparable to the knockdown cells was observed (C) ShRNA6 (27%), ShRNA6-T154A (20%), (D) ShRNA7 (23%) and ShRNA7-T154A (21%). The TrCx43 mutant produced migration levels similar to control cells. The experiment was repeated three times with Control (n = 61spheroids), ShRNA6 (n = 55 spheroids), ShRNA6-T154A (n = 58 spheroids), ShRNA6-TrCx43 (n = 61spheroids); Control (n = 62 spheroids), ShRNA7 (n = 61 spheroids), ShRNA7-T154A (n = 60 spheroids), and ShRNA7-TrCx43 (n = 58 spheroids). One way Anova followed by Dunn's Method to do pairwise multiple comparisons was used to calculate significance. *p < 0.05 was considered significant. Scale bar = 100 μm.
