Fig. 3.
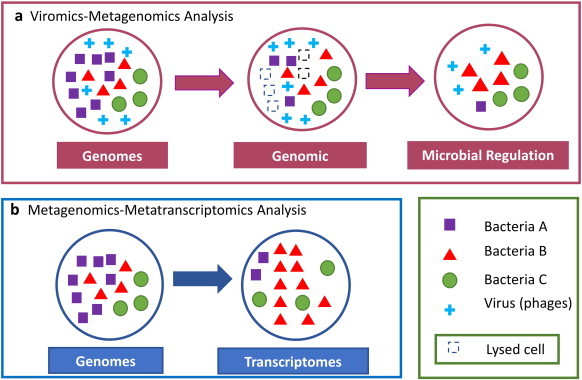
Molecular interactions explored using metagenomics–viromics and metagenomics–metatranscriptomics analyses. The interactions between microorganisms in the human microbiome can be better studied combining omics analysis. (a) In this panel is illustrated how phages can interact and affect the microbial diversity by infecting their host bacteria and thus promoting homeostasis or disbyosis [179,24]. This type of interaction can be explored using viromics combined with metagenomics. (b) The species abundance of the three hypothetical bacteria can be different depending on the used analysis (metagenomics or metagenomics combined with metatranscriptomics) [171]. The data integration and normalization when metagenomics is combined with metatranscriptomics is important because the metatranscriptomics data can correspond to different species abundance and/or to differentially expressed transcriptomes.
