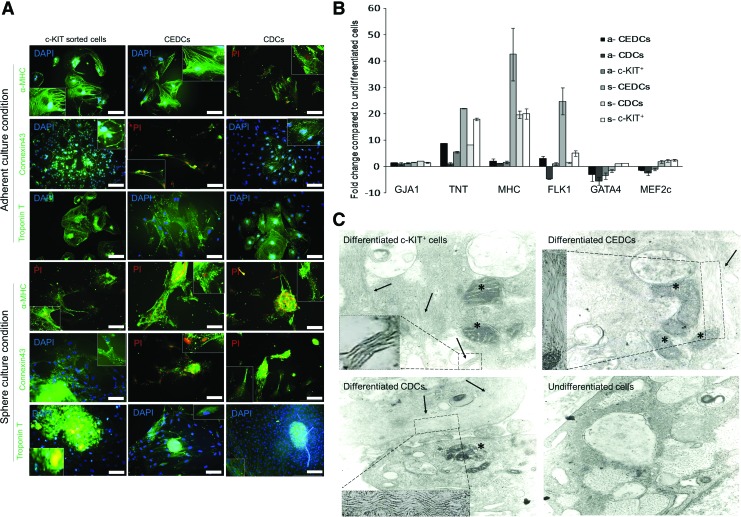
FIG. 4.
Characterization of cardiomyogenic differentiation efficiency. (A) Immune-staining was performed for three different CSCs subpopulations, which were differentiated into cardiomyocytes under two different culture conditions. cTnT, α-MHC and Connexin 43 were stained with fluorescent-conjugated antibodies (Scale bar=200 μm). (B) Real time reverse transcriptase PCR analyses for all six differentiation groups. Cardiac structural genes up-regulated after differentiation in all groups, whereas cardiac transcription factors downregulated in the adherent groups and upregulated in the sphere groups after myogenic differentiation. Flk-1 expression was upregulated in all groups except for adherently cultured CDCs. Expression of cardiac structural genes under the sphere condition was significantly higher than the adherent condition (P≤0.05). (C) Transmission electron microscopy results for isolated cells after differentiation. Myofibril polymers are shown with arrows (higher magnification of myofibril polymers are presented in dashes). Their arrangement is not obvious in the pictures. Of note, we observed large mitochondria (stars) in the cytoplasm of differentiated cells, which might be a good sign of cardiomyocyte differentiation of the cells. These changes are compared with undifferentiated control cells. Insets show higher magnifications of endoplasmic reticulum. Color images available online at www.liebertpub.com/scd