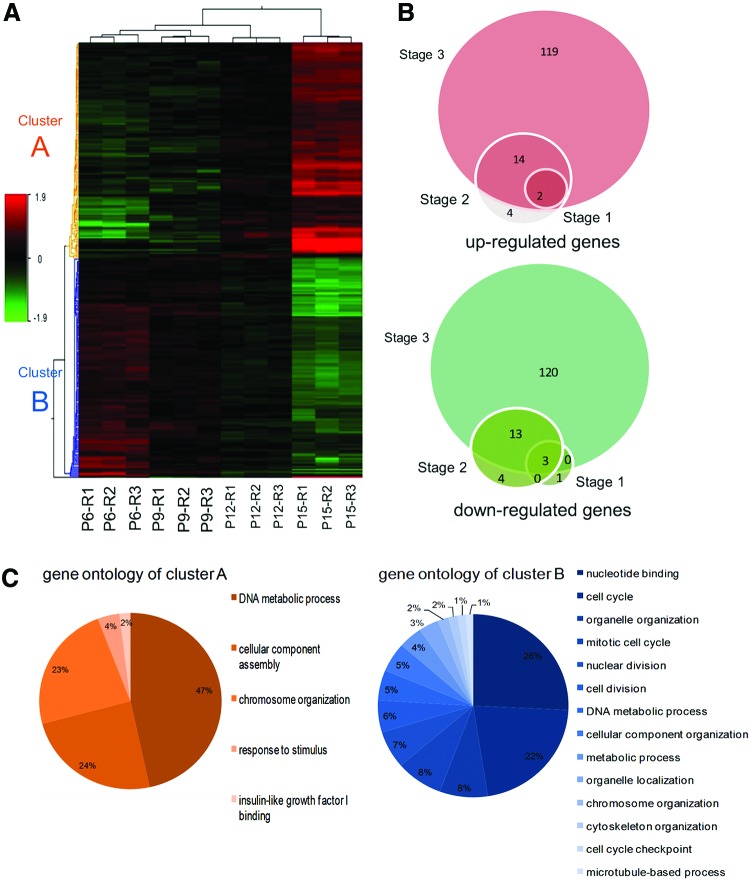
FIG. 5.
Whole genome expression analysis using microarray technology. (A) Hierarchical clustering of 280 differentially expressed genes was performed using the median signal intensity for each replicate. Three biological replicates of cells in passages 6, 9, 12 and 15 were compared and showed high intraclass correlations compared with interclass correlations. Two distinct clusters were distinguishable based on the expression patterns of cells, including transcripts that were up-regulated through passages that were highly expressed in passage 15 (cluster A) and transcripts downregulated by increasing the passages (cluster B). (B) Venn diagram showing the number of up- and downregulated genes in passages 9 (stage 1), 12 (stage 2) and 15 (stage 3) compared with passage 6. The number of significantly changed transcripts (P≤0.05, FC≥2) through subsequent passages is increased. (C) Functional classification of differentially expressed transcript was performed using BINGO software. It was shown that the main significant gene ontology of up-regulated transcripts (cluster A) was the DNA metabolic process while they were nucleotide binding and cell cycle for downregulated transcripts (cluster B). Color images available online at www.liebertpub.com/scd